Difference between revisions of "Nos"
| Line 1: | Line 1: | ||
| − | + | ||
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || production of nitric oxide | |style="background:#ABCDEF;" align="center"|'''Function''' || production of nitric oxide | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU07630 nos] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 38 kDa, 5.528 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 38 kDa, 5.528 | ||
Revision as of 15:55, 6 August 2012
| Gene name | nos |
| Synonyms | yflM |
| Essential | no |
| Product | nitric-oxide synthase |
| Function | production of nitric oxide |
| Gene expression levels in SubtiExpress: nos | |
| MW, pI | 38 kDa, 5.528 |
| Gene length, protein length | 1008 bp, 336 aa |
| Immediate neighbours | yflN, yflL |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 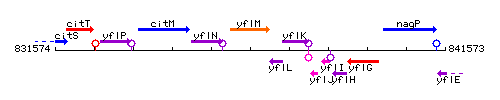
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
miscellaneous metabolic pathways, biosynthesis of antibacterial compounds
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU07630
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: Bacterial NOS oxygenase subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): heme PubMed
- Effectors of protein activity:
Database entries
- UniProt: O34453
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information: Note that the gene yflL is located between nos and yflK, but on the opposite strand.
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Isabelle Salard-Arnaud, Dennis Stuehr, Jean-Luc Boucher, Daniel Mansuy
Spectroscopic, catalytic and binding properties of Bacillus subtilis NO synthase-like protein: comparison with other bacterial and mammalian NO synthases.
J Inorg Biochem: 2012, 106(1);164-71
[PubMed:22119809]
[WorldCat.org]
[DOI]
(I p)
Frank Schreiber, Martin Beutler, Dennis Enning, María Lamprecht-Grandio, Olga Zafra, José Eduardo González-Pastor, Dirk de Beer
The role of nitric-oxide-synthase-derived nitric oxide in multicellular traits of Bacillus subtilis 3610: biofilm formation, swarming, and dispersal.
BMC Microbiol: 2011, 11;111
[PubMed:21599925]
[WorldCat.org]
[DOI]
(I e)
Zhi-Qiang Wang, Chin-Chuan Wei, Dennis J Stuehr
How does a valine residue that modulates heme-NO binding kinetics in inducible NO synthase regulate enzyme catalysis?
J Inorg Biochem: 2010, 104(3);349-56
[PubMed:20006999]
[WorldCat.org]
[DOI]
(I p)
Kartikeya Pant, Alexandrine M Bilwes, Subrata Adak, Dennis J Stuehr, Brian R Crane
Structure of a nitric oxide synthase heme protein from Bacillus subtilis.
Biochemistry: 2002, 41(37);11071-9
[PubMed:12220171]
[WorldCat.org]
[DOI]
(P p)
Subrata Adak, Kulwant S Aulak, Dennis J Stuehr
Direct evidence for nitric oxide production by a nitric-oxide synthase-like protein from Bacillus subtilis.
J Biol Chem: 2002, 277(18);16167-71
[PubMed:11856757]
[WorldCat.org]
[DOI]
(P p)