Difference between revisions of "LctP"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 48: | Line 48: | ||
* '''Locus tag:''' BSU03060 | * '''Locus tag:''' BSU03060 | ||
| − | |||
| − | |||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
Revision as of 17:59, 27 January 2012
- Description: lactate permease, excretion
| Gene name | lctP |
| Synonyms | ycgC |
| Essential | no |
| Product | L-lactate permease |
| Function | lactate excretion |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 57 kDa, 9.766 |
| Gene length, protein length | 1623 bp, 541 aa |
| Immediate neighbours | ldh, mdr |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 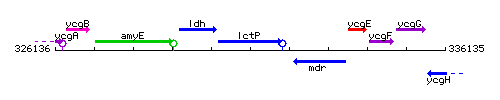
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
transporters/ other, carbon core metabolism, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU03060
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: lactate permease family (according to Swiss-Prot)
- Paralogous protein(s): LutP
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P55910
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
You-Kwan Oh, Bernhard O Palsson, Sung M Park, Christophe H Schilling, Radhakrishnan Mahadevan
Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data.
J Biol Chem: 2007, 282(39);28791-28799
[PubMed:17573341]
[WorldCat.org]
[DOI]
(P p)
Heike Reents, Richard Münch, Thorben Dammeyer, Dieter Jahn, Elisabeth Härtig
The Fnr regulon of Bacillus subtilis.
J Bacteriol: 2006, 188(3);1103-12
[PubMed:16428414]
[WorldCat.org]
[DOI]
(P p)
Jonas T Larsson, Annika Rogstam, Claes von Wachenfeldt
Coordinated patterns of cytochrome bd and lactate dehydrogenase expression in Bacillus subtilis.
Microbiology (Reading): 2005, 151(Pt 10);3323-3335
[PubMed:16207915]
[WorldCat.org]
[DOI]
(P p)
H Cruz Ramos, T Hoffmann, M Marino, H Nedjari, E Presecan-Siedel, O Dreesen, P Glaser, D Jahn
Fermentative metabolism of Bacillus subtilis: physiology and regulation of gene expression.
J Bacteriol: 2000, 182(11);3072-80
[PubMed:10809684]
[WorldCat.org]
[DOI]
(P p)