Difference between revisions of "MtlA"
(→Expression and regulation) |
|||
| Line 18: | Line 18: | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbohydrate_metabolic_pathways.html Sugar catabolism]''' | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbohydrate_metabolic_pathways.html Sugar catabolism]''' | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 50 kDa, 8.814 |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 1434 bp, 478 aa |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ycnL]]'', ''[[mtlF]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ycnL]]'', ''[[mtlF]]'' | ||
Revision as of 10:55, 8 November 2011
- Description: trigger enzyme: mannitol-specific phosphotransferase system, EIICB of the PTS
| Gene name | mtlA |
| Synonyms | |
| Essential | no |
| Product | trigger enzyme: mannitol-specific phosphotransferase system, EIICB |
| Function | mannitol uptake and phosphorylation, control of MtlR activity |
| Interactions involving this protein in SubtInteract: MtlA | |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 50 kDa, 8.814 |
| Gene length, protein length | 1434 bp, 478 aa |
| Immediate neighbours | ycnL, mtlF |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 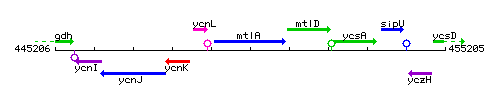
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
phosphotransferase systems, utilization of specific carbon sources, transcription factors and their control, trigger enzyme, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU03981
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Protein EIIA N(pi)-phospho-L-histidine + protein EIIB = protein EIIA + protein EIIB N(pi)-phospho-L-histidine/cysteine (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylation on Ser-559 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- UniProt: P42956
- KEGG entry: [3]
- E.C. number: 2.7.1.69
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Kambiz Morabbi Heravi, Marian Wenzel, Josef Altenbuchner
Regulation of mtl operon promoter of Bacillus subtilis: requirements of its use in expression vectors.
Microb Cell Fact: 2011, 10;83
[PubMed:22014119]
[WorldCat.org]
[DOI]
(I e)
Philippe Joyet, Meriem Derkaoui, Sandrine Poncet, Josef Deutscher
Control of Bacillus subtilis mtl operon expression by complex phosphorylation-dependent regulation of the transcriptional activator MtlR.
Mol Microbiol: 2010, 76(5);1279-94
[PubMed:20444094]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Shouji Watanabe, Miyuki Hamano, Hiroshi Kakeshita, Keigo Bunai, Shigeo Tojo, Hirotake Yamaguchi, Yasutaro Fujita, Sui-Lam Wong, Kunio Yamane
Mannitol-1-phosphate dehydrogenase (MtlD) is required for mannitol and glucitol assimilation in Bacillus subtilis: possible cooperation of mtl and gut operons.
J Bacteriol: 2003, 185(16);4816-24
[PubMed:12897001]
[WorldCat.org]
[DOI]
(P p)
Jonathan Reizer, Steffi Bachem, Aiala Reizer, Maryvonne Arnaud, Milton H Saier, Jörg Stülke
Novel phosphotransferase system genes revealed by genome analysis - the complete complement of PTS proteins encoded within the genome of Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 12);3419-3429
[PubMed:10627040]
[WorldCat.org]
[DOI]
(P p)