Difference between revisions of "PnpA"
(→Extended information on the protein) |
|||
| Line 81: | Line 81: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''[[Interactions]]:''' part of the [[RNA degradosome]] | + | * '''[[SubtInteract|Interactions]]:''' |
| − | ** [[PnpA]]-[[RnjA]] {{PubMed|19193632}} | + | ** part of the [[RNA degradosome]] {{PubMed|19193632}} |
| + | ** [[PnpA]]-[[RnjA]] {{PubMed|19193632}} | ||
| + | ** [[PnpA]]-[[PfkA]] {{PubMed|19193632}} | ||
| + | ** [[PnpA]]-[[Rny]] {{PubMed|19193632,21803996}} | ||
| + | ** [[PnpA]]-[[CshA]] {{PubMed|20572937}} | ||
| − | * '''Localization:''' | + | * '''[[Localization]]:''' |
=== Database entries === | === Database entries === | ||
Revision as of 09:37, 2 August 2011
- Description: polynucleotide phosphorylase, RNase
| Gene name | pnpA |
| Synonyms | comR |
| Essential | no |
| Product | polynucleotide phosphorylase (PNPase) (EC 2.7.7.8) |
| Function | necessary for competence development |
| Interactions involving this protein in SubtInteract: PnpA | |
| MW, pI | 77 kDa, 4.89 |
| Gene length, protein length | 2115 bp, 705 aa |
| Immediate neighbours | rpsO, ylxY |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 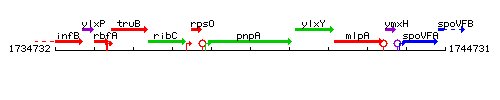
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU16690
Phenotypes of a mutant
- The pnpA mutant is cold sensitive and sensitive to tetracyclin, it shows multiseptate filamentous growth. PubMed
- The mutant is deficient in genetic competence (no expression of the late competence genes) PubMed
- The mutant overexpresses the trp and ycgM-ycgN-ycgO operons.
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 3'-5' exoribonuclease, RNase, PNPase degrades the trp mRNA from the RNA-TRAP complex
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 3CDI (protein from E. coli), 3GCM (protein from E. coli, PNPase/RNase E micro-domain/RNA tetragonal crystal form )
- UniProt: P50849
- KEGG entry: [2]
- E.C. number:
Additional information
required for the expression of late competence genes comGA and comK, requirement bypassed by a mecA disruption; may be necessary for modification of the srfAA transcript (stabilization or translation activation)
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP584 (aphA3), available in Stülke lab
- Expression vector:
- for expression, purification in E. coli with N-terminal His-tag, in pWH844: pGP838, available in Stülke lab
- for expression/ purification from B. subtilis with N-terminal Strep-tag, for SPINE, in pGP380: pGP1342, available in Stülke lab
- for chromosomal expression of PnpA-Strep (cat): GP1002, available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
David Bechhofer, Mount Sinai School, New York, USA Homepage
Your additional remarks
References
Reviews
José M Andrade, Vânia Pobre, Inês J Silva, Susana Domingues, Cecília M Arraiano
The role of 3'-5' exoribonucleases in RNA degradation.
Prog Mol Biol Transl Sci: 2009, 85;187-229
[PubMed:19215773]
[WorldCat.org]
[DOI]
(P p)
Sue Lin-Chao, Ni-Ting Chiou, Gadi Schuster
The PNPase, exosome and RNA helicases as the building components of evolutionarily-conserved RNA degradation machines.
J Biomed Sci: 2007, 14(4);523-32
[PubMed:17514363]
[WorldCat.org]
[DOI]
(P p)
Devanand Sarkar, Paul B Fisher
Polynucleotide phosphorylase: an evolutionary conserved gene with an expanding repertoire of functions.
Pharmacol Ther: 2006, 112(1);243-63
[PubMed:16733069]
[WorldCat.org]
[DOI]
(P p)
A J Carpousis, N F Vanzo, L C Raynal
mRNA degradation. A tale of poly(A) and multiprotein machines.
Trends Genet: 1999, 15(1);24-8
[PubMed:10087930]
[WorldCat.org]
[DOI]
(P p)
Original publications
Martin Lehnik-Habrink, Joseph Newman, Fabian M Rothe, Alexandra S Solovyova, Cecilia Rodrigues, Christina Herzberg, Fabian M Commichau, Richard J Lewis, Jörg Stülke
RNase Y in Bacillus subtilis: a Natively disordered protein that is the functional equivalent of RNase E from Escherichia coli.
J Bacteriol: 2011, 193(19);5431-41
[PubMed:21803996]
[WorldCat.org]
[DOI]
(I p)
Martin Lehnik-Habrink, Henrike Pförtner, Leonie Rempeters, Nico Pietack, Christina Herzberg, Jörg Stülke
The RNA degradosome in Bacillus subtilis: identification of CshA as the major RNA helicase in the multiprotein complex.
Mol Microbiol: 2010, 77(4);958-71
[PubMed:20572937]
[WorldCat.org]
[DOI]
(I p)
Shiyi Yao, David H Bechhofer
Initiation of decay of Bacillus subtilis rpsO mRNA by endoribonuclease RNase Y.
J Bacteriol: 2010, 192(13);3279-86
[PubMed:20418391]
[WorldCat.org]
[DOI]
(I p)
Juan Campos-Guillén, Jackeline Lizzeta Arvizu-Gómez, George H Jones, Gabriela Olmedo-Alvarez
Characterization of tRNA(Cys) processing in a conditional Bacillus subtilis CCase mutant reveals the participation of RNase R in its quality control.
Microbiology (Reading): 2010, 156(Pt 7);2102-2111
[PubMed:20360175]
[WorldCat.org]
[DOI]
(I p)
Gintaras Deikus, David H Bechhofer
Bacillus subtilis trp Leader RNA: RNase J1 endonuclease cleavage specificity and PNPase processing.
J Biol Chem: 2009, 284(39);26394-401
[PubMed:19638340]
[WorldCat.org]
[DOI]
(I p)
Shiyi Yao, David H Bechhofer
Processing and stability of inducibly expressed rpsO mRNA derivatives in Bacillus subtilis.
J Bacteriol: 2009, 191(18);5680-9
[PubMed:19633085]
[WorldCat.org]
[DOI]
(I p)
Paula P Cardenas, Begoña Carrasco, Humberto Sanchez, Gintaras Deikus, David H Bechhofer, Juan C Alonso
Bacillus subtilis polynucleotide phosphorylase 3'-to-5' DNase activity is involved in DNA repair.
Nucleic Acids Res: 2009, 37(12);4157-69
[PubMed:19433509]
[WorldCat.org]
[DOI]
(I p)
Fabian M Commichau, Fabian M Rothe, Christina Herzberg, Eva Wagner, Daniel Hellwig, Martin Lehnik-Habrink, Elke Hammer, Uwe Völker, Jörg Stülke
Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing.
Mol Cell Proteomics: 2009, 8(6);1350-60
[PubMed:19193632]
[WorldCat.org]
[DOI]
(I p)
Juan Campos-Guillén, Patricia Bralley, George H Jones, David H Bechhofer, Gabriela Olmedo-Alvarez
Addition of poly(A) and heteropolymeric 3' ends in Bacillus subtilis wild-type and polynucleotide phosphorylase-deficient strains.
J Bacteriol: 2005, 187(14);4698-706
[PubMed:15995184]
[WorldCat.org]
[DOI]
(P p)
Irina A Oussenko, Teppei Abe, Hiromi Ujiie, Akira Muto, David H Bechhofer
Participation of 3'-to-5' exoribonucleases in the turnover of Bacillus subtilis mRNA.
J Bacteriol: 2005, 187(8);2758-67
[PubMed:15805522]
[WorldCat.org]
[DOI]
(P p)
Gintaras Deikus, Paul Babitzke, David H Bechhofer
Recycling of a regulatory protein by degradation of the RNA to which it binds.
Proc Natl Acad Sci U S A: 2004, 101(9);2747-51
[PubMed:14976255]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Georg Homuth, Christian Scharf, Michael Hecker
Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis.
J Bacteriol: 2002, 184(9);2500-20
[PubMed:11948165]
[WorldCat.org]
[DOI]
(P p)
G A Farr, I A Oussenko, D H Bechhofer
Protection against 3'-to-5' RNA decay in Bacillus subtilis.
J Bacteriol: 1999, 181(23);7323-30
[PubMed:10572137]
[WorldCat.org]
[DOI]
(P p)
D H Bechhofer, W Wang
Decay of ermC mRNA in a polynucleotide phosphorylase mutant of Bacillus subtilis.
J Bacteriol: 1998, 180(22);5968-77
[PubMed:9811656]
[WorldCat.org]
[DOI]
(P p)
W Wang, D H Bechhofer
Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain.
J Bacteriol: 1996, 178(8);2375-82
[PubMed:8636041]
[WorldCat.org]
[DOI]
(P p)
A Luttinger, J Hahn, D Dubnau
Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis.
Mol Microbiol: 1996, 19(2);343-56
[PubMed:8825779]
[WorldCat.org]
[DOI]
(P p)
S Mitra, K Hue, D H Bechhofer
In vitro processing activity of Bacillus subtilis polynucleotide phosphorylase.
Mol Microbiol: 1996, 19(2);329-42
[PubMed:8825778]
[WorldCat.org]
[DOI]
(P p)
M P Deutscher, N B Reuven
Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis.
Proc Natl Acad Sci U S A: 1991, 88(8);3277-80
[PubMed:1707536]
[WorldCat.org]
[DOI]
(P p)
PNPase in E. coli