Difference between revisions of "RpoB"
(→Database entries) |
(→References) |
||
| Line 148: | Line 148: | ||
=References= | =References= | ||
| − | |||
| − | |||
'''Additional publications:''' {{PubMed|20702425,20817769}} | '''Additional publications:''' {{PubMed|20702425,20817769}} | ||
| + | <pubmed> 19680289 17644585 7657605,12486038,10675340,7592585, 20525796, 17981983, 18763711, 19191541 21512256</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 11:01, 23 April 2011
- Description: RNA polymerase beta subunit
| Gene name | rpoB |
| Synonyms | |
| Essential | yes PubMed |
| Product | RNA polymerase beta subunit |
| Function | transcription |
| MW, pI | 133 kDa, 4.731 |
| Gene length, protein length | 3579 bp, 1193 aa |
| Immediate neighbours | ybxB, rpoC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 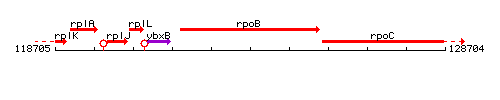
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
transcription, essential genes, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01070
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Nucleoside triphosphate + RNA(n) = diphosphate + RNA(n+1) (according to Swiss-Prot)
- Protein family: RNA polymerase beta chain family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- RpoA-RpoB-RpoC PubMed, NusA-RpoB PubMed
- SigA-(RpoB-RpoC) PubMed, SigB-(RpoB-RpoC)
- SigD-(RpoB-RpoC), SigE-(RpoB-RpoC)
- SigF-(RpoB-RpoC), SigG-(RpoB-RpoC)
- SigH-(RpoB-RpoC), SigI-(RpoB-RpoC)
- SigK-(RpoB-RpoC), SigL-(RpoB-RpoC)
- SigM-(RpoB-RpoC), SigV-(RpoB-RpoC)
- SigW-(RpoB-RpoC), SigX-(RpoB-RpoC)
- SigY-(RpoB-RpoC), SigZ-(RpoB-RpoC)
- Xpf-(RpoB-RpoC), YlaC-(RpoB-RpoC)
- YvrI-RpoB PubMed,
- Mfd-RpoB PubMed
- Localization: membrane associated PubMed
Database entries
- Structure:
- UniProt: P37870
- KEGG entry: [3]
- E.C. number: 2.7.7.6
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Operon: rpoB DBTBS
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Takashi Inaoka, Kozo Ochi
Activation of dormant secondary metabolism neotrehalosadiamine synthesis by an RNA polymerase mutation in Bacillus subtilis.
Biosci Biotechnol Biochem: 2011, 75(4);618-23
[PubMed:21512256]
[WorldCat.org]
[DOI]
(I p)
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Xiao Yang, Seeseei Molimau, Geoff P Doherty, Elecia B Johnston, Jon Marles-Wright, Rosalba Rothnagel, Ben Hankamer, Richard J Lewis, Peter J Lewis
The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA.
EMBO Rep: 2009, 10(9);997-1002
[PubMed:19680289]
[WorldCat.org]
[DOI]
(I p)
Amy E Perkins, Andrew C Schuerger, Wayne L Nicholson
Isolation of rpoB mutations causing rifampicin resistance in Bacillus subtilis spores exposed to simulated Martian surface conditions.
Astrobiology: 2008, 8(6);1159-67
[PubMed:19191541]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Ulf Gerth, Holger Kock, Ilja Kusters, Stephan Michalik, Robert L Switzer, Michael Hecker
Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis.
J Bacteriol: 2008, 190(1);321-31
[PubMed:17981983]
[WorldCat.org]
[DOI]
(I p)
Amy E Perkins, Wayne L Nicholson
Uncovering new metabolic capabilities of Bacillus subtilis using phenotype profiling of rifampin-resistant rpoB mutants.
J Bacteriol: 2008, 190(3);807-14
[PubMed:17644585]
[WorldCat.org]
[DOI]
(I p)
Claudia Rollenhagen, Haike Antelmann, Janine Kirstein, Olivier Delumeau, Michael Hecker, Michael D Yudkin
Binding of sigma(A) and sigma(B) to core RNA polymerase after environmental stress in Bacillus subtilis.
J Bacteriol: 2003, 185(1);35-40
[PubMed:12486038]
[WorldCat.org]
[DOI]
(P p)
P J Lewis, S D Thaker, J Errington
Compartmentalization of transcription and translation in Bacillus subtilis.
EMBO J: 2000, 19(4);710-8
[PubMed:10675340]
[WorldCat.org]
[DOI]
(P p)
X Yang, C W Price
Streptolydigin resistance can be conferred by alterations to either the beta or beta' subunits of Bacillus subtilis RNA polymerase.
J Biol Chem: 1995, 270(41);23930-3
[PubMed:7592585]
[WorldCat.org]
[DOI]
(P p)
K J Boor, M L Duncan, C W Price
Genetic and transcriptional organization of the region encoding the beta subunit of Bacillus subtilis RNA polymerase.
J Biol Chem: 1995, 270(35);20329-36
[PubMed:7657605]
[WorldCat.org]
[DOI]
(P p)