CheY
- Description: two-component response regulator, modulation of flagellar switch bias
| Gene name | cheY |
| Synonyms | cheB |
| Essential | no |
| Product | two-component response regulator |
| Function | modulation of flagellar switch bias |
| Gene expression levels in SubtiExpress: cheY | |
| Interactions involving this protein in SubtInteract: CheY | |
| MW, pI | 13 kDa, 4.746 |
| Gene length, protein length | 360 bp, 120 aa |
| Immediate neighbours | fliY, fliZ |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 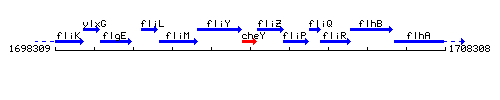
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed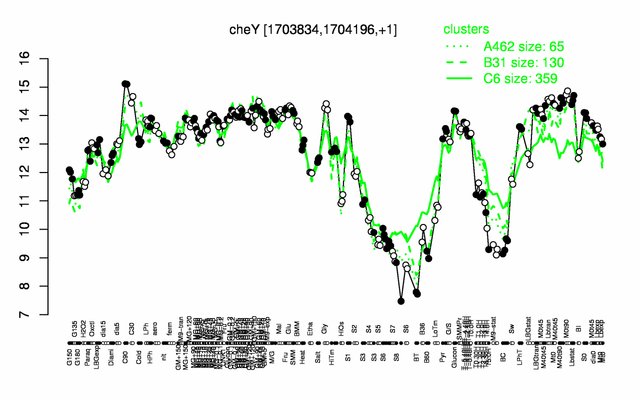
| |
Contents
Categories containing this gene/protein
transcription factors and their control, motility and chemotaxis, phosphoproteins
This gene is a member of the following regulons
CodY regulon, DegU regulon, SigD regulon, Spo0A regulon
The gene
Basic information
- Locus tag: BSU16330
Phenotypes of a mutant
- not essential for pellicle biofilm formation, but mutant is outcompeted by the wild-type strain when competed during pellicle formation PubMed
Database entries
- BsubCyc: BSU16330
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): YneI
Extended information on the protein
- Kinetic information:
- Effectors of protein activity:
- Localization:
- cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU16330
- UniProt: P24072
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: part of the fla-che operon
- Regulation: see fla-che operon
- Regulatory mechanism: see fla-che operon
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original Publications
Serena Mordini, Cecilia Osera, Simone Marini, Francesco Scavone, Riccardo Bellazzi, Alessandro Galizzi, Cinzia Calvio
The role of SwrA, DegU and P(D3) in fla/che expression in B. subtilis.
PLoS One: 2013, 8(12);e85065
[PubMed:24386445]
[WorldCat.org]
[DOI]
(I e)
Vincent J Cannistraro, George D Glekas, Christopher V Rao, George W Ordal
Cellular stoichiometry of the chemotaxis proteins in Bacillus subtilis.
J Bacteriol: 2011, 193(13);3220-7
[PubMed:21515776]
[WorldCat.org]
[DOI]
(I p)
Y Pazy, M A Motaleb, M T Guarnieri, N W Charon, R Zhao, R E Silversmith
Identical phosphatase mechanisms achieved through distinct modes of binding phosphoprotein substrate.
Proc Natl Acad Sci U S A: 2010, 107(5);1924-9
[PubMed:20080618]
[WorldCat.org]
[DOI]
(I p)
Travis J Muff, George W Ordal
The CheC phosphatase regulates chemotactic adaptation through CheD.
J Biol Chem: 2007, 282(47);34120-8
[PubMed:17908686]
[WorldCat.org]
[DOI]
(P p)
Kazuo Kobayashi
Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis.
Mol Microbiol: 2007, 66(2);395-409
[PubMed:17850253]
[WorldCat.org]
[DOI]
(P p)
Travis J Muff, Richard M Foster, Peter J Y Liu, George W Ordal
CheX in the three-phosphatase system of bacterial chemotaxis.
J Bacteriol: 2007, 189(19);7007-13
[PubMed:17675386]
[WorldCat.org]
[DOI]
(P p)
H Werhane, P Lopez, M Mendel, M Zimmer, G W Ordal, L M Márquez-Magaña
The last gene of the fla/che operon in Bacillus subtilis, ylxL, is required for maximal sigmaD function.
J Bacteriol: 2004, 186(12);4025-9
[PubMed:15175317]
[WorldCat.org]
[DOI]
(P p)
Hendrik Szurmant, Travis J Muff, George W Ordal
Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade.
J Biol Chem: 2004, 279(21);21787-92
[PubMed:14749334]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
Hendrik Szurmant, Michael W Bunn, Vincent J Cannistraro, George W Ordal
Bacillus subtilis hydrolyzes CheY-P at the location of its action, the flagellar switch.
J Biol Chem: 2003, 278(49);48611-6
[PubMed:12920116]
[WorldCat.org]
[DOI]
(P p)
J R Kirby, M M Saulmon, C J Kristich, G W Ordal
CheY-dependent methylation of the asparagine receptor, McpB, during chemotaxis in Bacillus subtilis.
J Biol Chem: 1999, 274(16);11092-100
[PubMed:10196193]
[WorldCat.org]
[DOI]
(P p)
W Estacio, S S Anna-Arriola, M Adedipe, L M Márquez-Magaña
Dual promoters are responsible for transcription initiation of the fla/che operon in Bacillus subtilis.
J Bacteriol: 1998, 180(14);3548-55
[PubMed:9657996]
[WorldCat.org]
[DOI]
(P p)
J R Kirby, C J Kristich, S L Feinberg, G W Ordal
Methanol production during chemotaxis to amino acids in Bacillus subtilis.
Mol Microbiol: 1997, 24(4);869-78
[PubMed:9194713]
[WorldCat.org]
[DOI]
(P p)
L M Márquez-Magaña, M J Chamberlin
Characterization of the sigD transcription unit of Bacillus subtilis.
J Bacteriol: 1994, 176(8);2427-34
[PubMed:8157612]
[WorldCat.org]
[DOI]
(P p)
D S Bischoff, R B Bourret, M L Kirsch, G W Ordal
Purification and characterization of Bacillus subtilis CheY.
Biochemistry: 1993, 32(35);9256-61
[PubMed:8369293]
[WorldCat.org]
[DOI]
(P p)