Difference between revisions of "SftA"
(→Reviews) |
|||
| Line 144: | Line 144: | ||
=References= | =References= | ||
==Reviews== | ==Reviews== | ||
| − | <pubmed> 22047950 </pubmed> | + | <pubmed> 22047950 22934648 </pubmed> |
| + | |||
==Original publications== | ==Original publications== | ||
<pubmed>16479537 19788545 19818024 21239579</pubmed> | <pubmed>16479537 19788545 19818024 21239579</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 17:07, 29 July 2013
- Description: DNA translocase
| Gene name | sftA |
| Synonyms | ytpS, cdtA |
| Essential | no |
| Product | DNA translocase |
| Function | resolution of chromosome dimers |
| Gene expression levels in SubtiExpress: sftA | |
| MW, pI | 107 kDa, 5.052 |
| Gene length, protein length | 2856 bp, 952 aa |
| Immediate neighbours | murC, ytpR |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 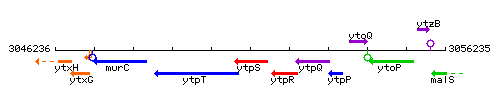
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29805
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- aids in moving DNA away from the closing septum, binds DNA, DNA-dependent ATPase activity PubMed
- the two DNA translocases SftA and SpoIIIE synergistically affect chromosome dimer resolution presumably by positioning the dif sites in close proximity, before or after completion of cell division, respectively PubMed
- Protein family:
- Paralogous protein(s): SpoIIIE
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- forms a hexamer PubMed
Database entries
- Structure:
- UniProt: C0SP86
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Rodrigo Reyes-Lamothe, Emilien Nicolas, David J Sherratt
Chromosome replication and segregation in bacteria.
Annu Rev Genet: 2012, 46;121-43
[PubMed:22934648]
[WorldCat.org]
[DOI]
(I p)
Christine Kaimer, Peter L Graumann
Players between the worlds: multifunctional DNA translocases.
Curr Opin Microbiol: 2011, 14(6);719-25
[PubMed:22047950]
[WorldCat.org]
[DOI]
(I p)
Original publications
Christine Kaimer, Katrin Schenk, Peter L Graumann
Two DNA translocases synergistically affect chromosome dimer resolution in Bacillus subtilis.
J Bacteriol: 2011, 193(6);1334-40
[PubMed:21239579]
[WorldCat.org]
[DOI]
(I p)
Christine Kaimer, José Eduardo González-Pastor, Peter L Graumann
SpoIIIE and a novel type of DNA translocase, SftA, couple chromosome segregation with cell division in Bacillus subtilis.
Mol Microbiol: 2009, 74(4);810-25
[PubMed:19818024]
[WorldCat.org]
[DOI]
(I p)
Steven J Biller, William F Burkholder
The Bacillus subtilis SftA (YtpS) and SpoIIIE DNA translocases play distinct roles in growing cells to ensure faithful chromosome partitioning.
Mol Microbiol: 2009, 74(4);790-809
[PubMed:19788545]
[WorldCat.org]
[DOI]
(I p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)