Difference between revisions of "FadF"
| Line 37: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/> | ||
| Line 118: | Line 114: | ||
** repressed in the absence of long-chain fatty acids ([[FadR]]) {{PubMed|17189250}} | ** repressed in the absence of long-chain fatty acids ([[FadR]]) {{PubMed|17189250}} | ||
** repressed by glucose (5-fold) [http://www.ncbi.nlm.nih.gov/pubmed/12850135 PubMed] | ** repressed by glucose (5-fold) [http://www.ncbi.nlm.nih.gov/pubmed/12850135 PubMed] | ||
| + | ** strongly induced in response to glucose starvation in M9 medium {{PubMed|23033921}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| Line 143: | Line 140: | ||
=References= | =References= | ||
| − | + | '''Additional publications:''' {{PubMed|23033921}} | |
<pubmed>12850135, 17189250</pubmed> | <pubmed>12850135, 17189250</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 19:09, 8 October 2012
- Description: similar to iron-sulphur-binding reductase
| Gene name | fadF |
| Synonyms | ywjF |
| Essential | no |
| Product | unknown |
| Function | fatty acid degradation |
| Gene expression levels in SubtiExpress: fadF | |
| Metabolic function and regulation of this protein in SubtiPathways: Fatty acid degradation | |
| MW, pI | 79 kDa, 6.526 |
| Gene length, protein length | 2115 bp, 705 aa |
| Immediate neighbours | acdA, ywjE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 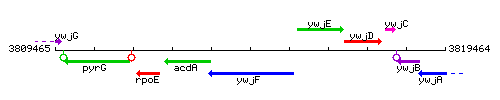
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed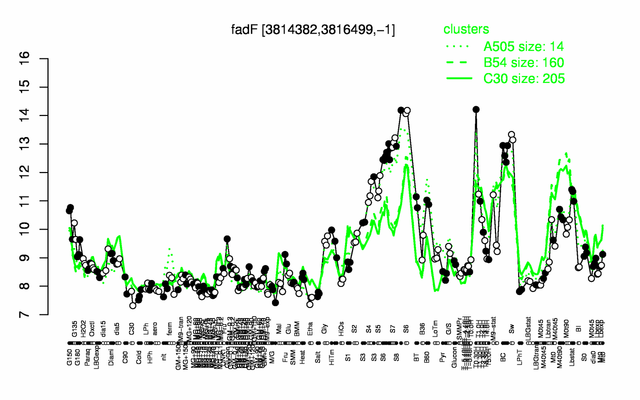
| |
Contents
Categories containing this gene/protein
utilization of lipids, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU37180
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): contains an iron-sulfur cluster
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P45866
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Hiroshi Matsuoka, Kazutake Hirooka, Yasutaro Fujita
Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation.
J Biol Chem: 2007, 282(8);5180-94
[PubMed:17189250]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)