Hbs
- Description: non-specific DNA-binding protein Hbsu
| Gene name | hbs |
| Synonyms | dbpA, hupA |
| Essential | yes PubMed |
| Product | non-specific DNA-binding protein Hbsu |
| Function | DNA packaging, function of the signal recognition complex |
| Gene expression levels in SubtiExpress: hbs | |
| MW, pI | 9 kDa, 9.501 |
| Gene length, protein length | 276 bp, 92 aa |
| Immediate neighbours | folE, spoIVA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 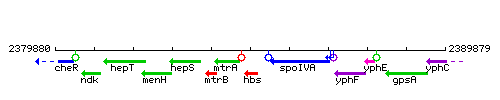
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
DNA condensation/ segregation, essential genes, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22790
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU22790
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: DbpA subfamily (according to Swiss-Prot)
- Paralogous protein(s): YonN
Extended information on the protein
- Kinetic information:
- Domains:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU22790
- Structure: 1HUU (Geobacillus stearothermophilus)
- UniProt: P08821
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: hbs PubMed
- Regulation:
- Additional information:
- mRNA is protected from degradation by RnjA by initiating ribosomes PubMed
- Hbs is present with about 50,000 copies per genome in vegetative cells and spores, it is one of the most abundant proteins in B. subtilis PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 19917 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 41521 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Fabian Commichau's lab
- Antibody:
Labs working on this gene/protein
- Ciaran Condon, IBPC, Paris, France Homepage
- Mohamed Marahiel, University of Marburg, Germany Homepage
Your additional remarks
References
Reviews
Original publications