DivIVA
- Description: curvature sensitive membrane binding protein that recruits other proteins to the poles and the division septum, cell-division initiation protein (septum placement), part of the Min system (with MinC, MinD, MinJ), Noc and the Min system ensure the efficient utilization of the division site at midcell in by ensuring Z ring placement
| Gene name | divIVA |
| Synonyms | ylmJ |
| Essential | no |
| Product | cell-division initiation protein |
| Function | septum placement |
| Gene expression levels in SubtiExpress: divIVA | |
| Interactions involving this protein in SubtInteract: DivIVA | |
| MW, pI | 19 kDa, 4.846 |
| Gene length, protein length | 492 bp, 164 aa |
| Immediate neighbours | ylmH, ileS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 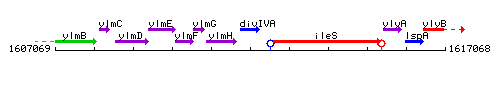
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell division, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU15420
Phenotypes of a mutant
- Deletion of divIVA leads to filamentation and polar divisions that in turn cause a minicell phenotype. PubMed
- A divIVA mutant has a severe sporulation defect. PubMed
Database entries
- BsubCyc: BSU15420
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
Filamentation is suppressed by mutations in minCD PubMed.
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- curvature sensitive membrane binding protein that recruits other proteins to the poles and the division septum
- DivIVA is required for polar localisation of MinC-MinD via MinJ. PubMed
- It also recruits RacA to the distal pole of the prespore PubMed.
- DivIVA may anchor SpoIIE briefly to the assembling polar septum before SpoIIE is subsequently released into the forespore membrane and recaptured at the polar septum PubMed
- required for the compartment-specific activation of SigF PubMed
- Protein family: gpsB family (according to Swiss-Prot)
- Paralogous protein(s): GpsB
Extended information on the protein
- Kinetic information:
- Modification:
- Cofactors: not known
- Effectors of protein activity: not known
- Localization:
- DivIVA forms a ring underneath the invaginating membrane at the site of cell division and is enriched at both cell poles PubMed.
- forms rings at the division septum and patches at the cell poles PubMed
- membrane targeting requires SecA PubMed
- assembles into a ring-like structure at the polar septum during sporulation PubMed
Database entries
- BsubCyc: BSU15420
- UniProt: P71021
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: divIVA PubMed
- Regulation:
- Additional information:
Biological materials
- Mutant:
- 4041 (divIVA::tet), available in Leendert Hamoen's, Jörg Stülke's, and Sven Halbedel 's lab
- GP1482 (chromosomal divIVA-Strep fusion, aphA3), purification from B. subtilis, for SPINE, available in Jörg Stülke's lab
- Expression vector:
- DivIVA-Strep available here
- pGP1497 (N-terminal Strep-tag fused to C-terminus of divIVA, TEV-site, purification from E. coli, in pGP172), available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion: divIVA-gfp fusions available from the Hamoen Lab
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Sven Halbedel's and Jörg Stülke's labs
- Antibody: A polyclonal anti-DivIVA antiserum generated in rabbit is described here PubMed.
Labs working on this gene/protein
Leendert Hamoen, Centre for Bacterial Cell Biology, Newcastle upon Tyne, United Kingdom [4]
Imrich Barak, Slovak Academy of Science, Bratislava, Slovakia homepage
Sven Halbedel, Robert Koch Institute homepage
Your additional remarks
References
Reviews
Henrik Strahl, Leendert W Hamoen
Finding the corners in a cell.
Curr Opin Microbiol: 2012, 15(6);731-6
[PubMed:23182676]
[WorldCat.org]
[DOI]
(I p)
Karan Gautam Kaval, Sven Halbedel
Architecturally the same, but playing a different game: the diverse species-specific roles of DivIVA proteins.
Virulence: 2012, 3(4);406-7
[PubMed:22722244]
[WorldCat.org]
[DOI]
(I p)
Marc Bramkamp, Suey van Baarle
Division site selection in rod-shaped bacteria.
Curr Opin Microbiol: 2009, 12(6);683-8
[PubMed:19884039]
[WorldCat.org]
[DOI]
(I p)
Jennifer R Juarez, William Margolin
Irresistible curves.
EMBO J: 2009, 28(15);2147-8
[PubMed:19654604]
[WorldCat.org]
[DOI]
(I p)
Original Publications