RecU
- Description: Holliday junction resolvase, DNA repair, homologous recombination and chromosome segregation; recognizes, distorts, and cleaves four-stranded recombination intermediates and modulates RecA activities, equivalent to E. coli RecU
| Gene name | recU |
| Synonyms | recG, prfA, yppB, jopB |
| Essential | no |
| Product | Holliday junction resolvase |
| Function | DNA repair/ recombination and chromosome segregation |
| Gene expression levels in SubtiExpress: recU | |
| Interactions involving this protein in SubtInteract: RecU | |
| MW, pI | 23 kDa, 9.511 |
| Gene length, protein length | 618 bp, 206 aa |
| Immediate neighbours | yppC, ponA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 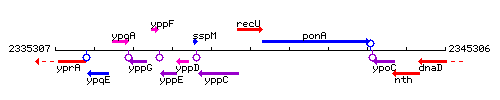
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
DNA repair/ recombination, cell envelope stress proteins (controlled by SigM, V, W, X, Y)
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22310
Phenotypes of a mutant
Database entries
- BsubCyc: BSU22310
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU22310
- Structure: 1ZP7
- UniProt: P39792
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- GP891 (recU::cat), available in Jörg Stülke's lab
- 1A895 (no resistance), PubMed, available at BGSC
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
RecU in other organisms
Ana R Pereira, Patricia Reed, Helena Veiga, Mariana G Pinho
The Holliday junction resolvase RecU is required for chromosome segregation and DNA damage repair in Staphylococcus aureus.
BMC Microbiol: 2013, 13;18
[PubMed:23356868]
[WorldCat.org]
[DOI]
(I e)