FabHA
Revision as of 14:06, 17 April 2014 by 134.76.70.252 (talk)
- Description: beta-ketoacyl-acyl carrier protein synthase III, principal condensing enzyme responsible for the initiation of fatty acid synthesis in non-stressed B. subtilis cells
| Gene name | fabHA |
| Synonyms | yjaX , fabH1 |
| Essential | no |
| Product | beta-ketoacyl-acyl carrier protein synthase III |
| Function | fatty acid biosynthesis |
| Gene expression levels in SubtiExpress: fabHA | |
| Metabolic function and regulation of this protein in SubtiPathways: fabHA | |
| MW, pI | 33 kDa, 5.045 |
| Gene length, protein length | 936 bp, 312 aa |
| Immediate neighbours | yjzB, fabF |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 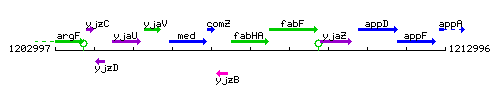
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU11330
Phenotypes of a mutant
- significant increase in the proportion of straight-chain fatty acids with a concomitant increase in 31:0-carbon phosphatidylethanolamine species PubMed
Database entries
- BsubCyc: BSU11330
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Acetyl-CoA + malonyl-[acyl-carrier-protein] = acetoacyl-[acyl-carrier-protein] + CoA + CO2 (according to Swiss-Prot)
- Protein family: fabH family (according to Swiss-Prot)
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization:
- cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU11330
- UniProt: O34746
- KEGG entry: [3]
Additional information
- affinity for butyryl-CoA, but prefers acetyl-CoA in fatty acid biosynthesis PubMed
Expression and regulation
- Regulation:
- expressed when the cells experience a lack of malonyl-CoA (FapR) PubMed
- inhibited by cerulenin PubMed
- induced upon fatty acid biosynthesis inhibition PubMed
- expression is reduced when SigW is activated (by alkaline shock, polymyxin B, vancomycin, cephalosporin C, D-cycloserine, and triton X-100) PubMed
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium): 897 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 1078 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 2129 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 1833 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 2586 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original Publications