CshA
Revision as of 13:01, 2 April 2014 by 134.76.38.147 (talk)
- Description: DEAD-box RNA helicase, important for adaptation to low temperatures
| Gene name | cshA |
| Synonyms | ydbR |
| Essential | no |
| Product | DEAD-box RNA helicase |
| Function | RNA helicase |
| Gene expression levels in SubtiExpress: cshA | |
| Interactions involving this protein in SubtInteract: CshA | |
| MW, pI | 57 kDa, 9.89 |
| Gene length, protein length | 1533 bp, 511 aa |
| Immediate neighbours | murF, ydbS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 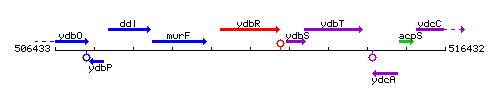
This image was kindly provided by SubtiList
| |
[None Expression at a glance] PubMed
| |
Contents
Categories containing this gene/protein
DEAD-box RNA helicases, translation, cold stress proteins, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU04580
Phenotypes of a mutant
- poor growth at low temperatures (16 to 20°C) PubMed
- reduced number of ribosomes PubMed
- no expression of the frlB-frlO-frlN-frlM-frlD operon PubMed
- strongly increased expression of the ysbA-ysbB operon PubMed
- transcription profile resulting from rny depletion: GEO PubMed
Database entries
- BsubCyc: BSU04580
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: helicase C-terminal domain (according to Swiss-Prot) DEAD-box RNA helicase
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasma, colocalizes with the ribosomes PubMed, cell membrane PubMed
Database entries
- BsubCyc: BSU04580
- Structure:
- UniProt: P96614
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- GP1035 (aphA3), available in Jörg Stülke's lab
- GP1083 (cat), available in Jörg Stülke's lab
- Expression vector:
- for expression/ purification from B. subtilis with C-terminal Strep-tag, for SPINE, in pGP382: pGP1387, available in Jörg Stülke's lab
- for expression/ purification from B. subtilis with C-terminal Strep-tag, for SPINE, expression from the native chromomsomal site: GP1026 (aphA3), available in Jörg Stülke's lab
- for expression/ purification from E. coli with N-terminal His-tag, in pWH844: pGP1386, available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- pGP1369 for chromosomal expression of CshA-YFP, available in Jörg Stülke's lab
- B. subtilis GP1081 cshA-gfp spc, available in Jörg Stülke's lab,
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- FLAG-tag construct:
- GP1010 (spc, based on pGP1331), available in Jörg Stülke's lab
- GP1074 (tet), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Mohamed Marahiel, Marburg University, Germany homepage
Your additional remarks
References
CshA in other organisms