RpmC
Revision as of 12:49, 2 April 2014 by 134.76.38.147 (talk)
- Description: ribosomal protein
| Gene name | rpmC |
| Synonyms | |
| Essential | no PubMed |
| Product | ribosomal protein L29 |
| Function | translation |
| Gene expression levels in SubtiExpress: rpmC | |
| Interactions involving this protein in SubtInteract: RpmC | |
| MW, pI | 7 kDa, 10.628 |
| Gene length, protein length | 198 bp, 66 aa |
| Immediate neighbours | rplP, rpsQ |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 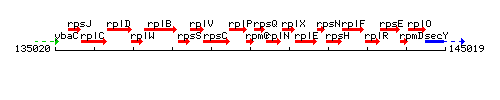
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed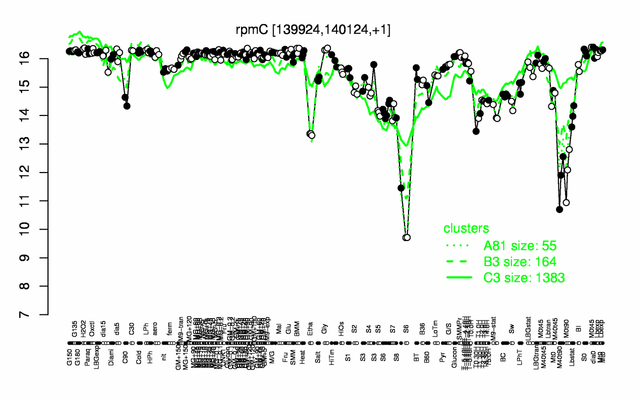
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01240
Phenotypes of a mutant
Database entries
- BsubCyc: BSU01240
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ribosomal protein L29P family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU01240
- Structure:
- UniProt: P12873
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon: rpsJ-rplC-rplD-rplW-rplB-rpsS-rplV-rpsC-rplP-rpmC-rpsQ-rplN-rplX-rplE-rpsN-rpsH-rplF-rplR-rpsE-rpmD-rplO PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References