IspD
- Description: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, third step in the MEP pathway of isoprenoid biosynthesis
| Gene name | ispD |
| Synonyms | yacM |
| Essential | yes PubMed |
| Product | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
| Function | MEP pathway of isoprenoid biosynthesis |
| Gene expression levels in SubtiExpress: ispD | |
| MW, pI | 25 kDa, 5.579 |
| Gene length, protein length | 696 bp, 232 aa |
| Immediate neighbours | yacL, ispF |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 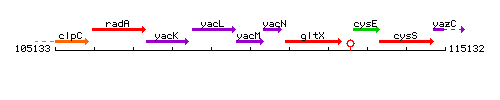
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed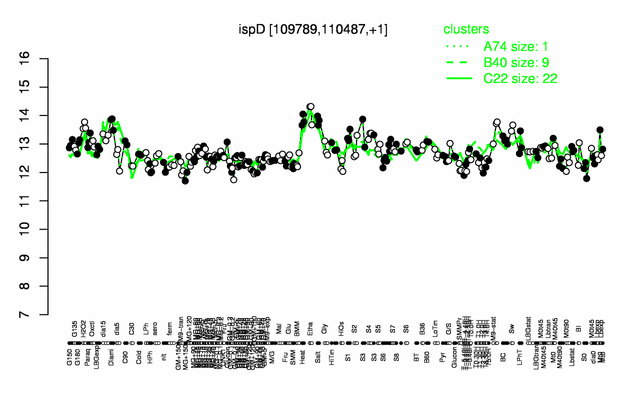
| |
Contents
Categories containing this gene/protein
biosynthesis of lipids, general stress proteins (controlled by SigB), cell envelope stress proteins (controlled by SigM, V, W, X, Y), essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU00900
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU00900
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: CTP + 2-C-methyl-D-erythritol 4-phosphate = diphosphate + 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol (according to Swiss-Prot)
- Protein family: ispD family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU00900
- UniProt: Q06755
- KEGG entry: [3]
- E.C. number: 2.7.7.60
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Becky M Hess, Junfeng Xue, Lye Meng Markillie, Ronald C Taylor, H Steven Wiley, Birgitte K Ahring, Bryan Linggi
Coregulation of Terpenoid Pathway Genes and Prediction of Isoprene Production in Bacillus subtilis Using Transcriptomics.
PLoS One: 2013, 8(6);e66104
[PubMed:23840410]
[WorldCat.org]
[DOI]
(I e)
Matthias C Witschel, H Wolfgang Höffken, Michael Seet, Liliana Parra, Thomas Mietzner, Frank Thater, Ricarda Niggeweg, Franz Röhl, Boris Illarionov, Felix Rohdich, Johannes Kaiser, Markus Fischer, Adelbert Bacher, François Diederich
Inhibitors of the herbicidal target IspD: allosteric site binding.
Angew Chem Int Ed Engl: 2011, 50(34);7931-5
[PubMed:21766403]
[WorldCat.org]
[DOI]
(I p)
Mattijs K Julsing, Michael Rijpkema, Herman J Woerdenbag, Wim J Quax, Oliver Kayser
Functional analysis of genes involved in the biosynthesis of isoprene in Bacillus subtilis.
Appl Microbiol Biotechnol: 2007, 75(6);1377-84
[PubMed:17458547]
[WorldCat.org]
[DOI]
(P p)
Adrian J Jervis, Penny D Thackray, Chris W Houston, Malcolm J Horsburgh, Anne Moir
SigM-responsive genes of Bacillus subtilis and their promoters.
J Bacteriol: 2007, 189(12);4534-8
[PubMed:17434969]
[WorldCat.org]
[DOI]
(P p)
Dirk Höper, Uwe Völker, Michael Hecker
Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis.
J Bacteriol: 2005, 187(8);2810-26
[PubMed:15805528]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, J Bernhardt, U Gerth, D Höper, T Koburger, U Völker, M Hecker
Identification of sigma(B)-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization.
J Bacteriol: 1999, 181(18);5718-24
[PubMed:10482513]
[WorldCat.org]
[DOI]
(P p)