RapJ
Revision as of 12:29, 16 May 2013 by 134.76.70.252 (talk)
- Description: response regulator aspartate phosphatase, dephosphorylates Spo0F-P
| Gene name | rapJ |
| Synonyms | ycdE |
| Essential | no |
| Product | response regulator aspartate phosphatase |
| Function | control of the phosphorelay |
| Gene expression levels in SubtiExpress: rapJ | |
| Interactions involving this protein in SubtInteract: RapJ | |
| MW, pI | 44 kDa, 4.902 |
| Gene length, protein length | 1119 bp, 373 aa |
| Immediate neighbours | cwlK, ycdF |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 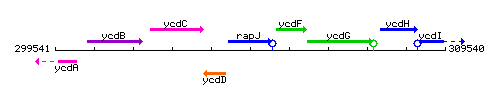
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
protein modification, phosphorelay, transcription factors and their control
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU02820
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Protein family: RAP family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- RapJ is made up of the C-terminal tetratricopeptide repeat (TPR) domain that is connected by a flexible helix containing linker to the N-terminal 3-helix bundle. Upon binding of the regulating peptide PhrC, the 3-helix bundle and the linker helix undergo a conformational change to form a TPR-like fold that merges with the existing C-terminal TPR domain. PubMed
- Modification:
- Cofactor(s):
Database entries
- UniProt: O34327
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: rapJ (according to DBTBS)
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References