YkfB
- Description: L-Ala-D/L-Glu epimerase
| Gene name | ykfB |
| Synonyms | |
| Essential | no |
| Product | L-Ala-D/L-Glu epimerase |
| Function | cell wall metabolism |
| Gene expression levels in SubtiExpress: ykfB | |
| MW, pI | 39 kDa, 5.905 |
| Gene length, protein length | 1098 bp, 366 aa |
| Immediate neighbours | ykfA, ykfC |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 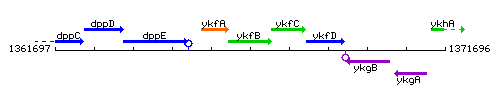
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed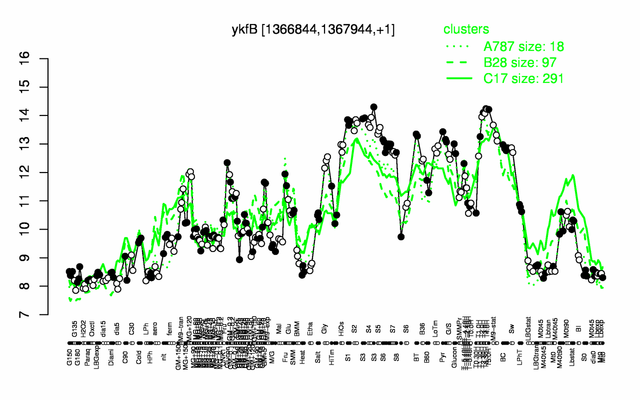
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU12980
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: O34508
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Tiit Lukk, Ayano Sakai, Chakrapani Kalyanaraman, Shoshana D Brown, Heidi J Imker, Ling Song, Alexander A Fedorov, Elena V Fedorov, Rafael Toro, Brandan Hillerich, Ronald Seidel, Yury Patskovsky, Matthew W Vetting, Satish K Nair, Patricia C Babbitt, Steven C Almo, John A Gerlt, Matthew P Jacobson
Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc Natl Acad Sci U S A: 2012, 109(11);4122-7
[PubMed:22392983]
[WorldCat.org]
[DOI]
(I p)
Vadim A Klenchin, Dawn M Schmidt, John A Gerlt, Ivan Rayment
Evolution of enzymatic activities in the enolase superfamily: structure of a substrate-liganded complex of the L-Ala-D/L-Glu epimerase from Bacillus subtilis.
Biochemistry: 2004, 43(32);10370-8
[PubMed:15301535]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Yoshiko Nakaura, Robert P Shivers, Hirotake Yamaguchi, Richard Losick, Yasutaro Fujita, Abraham L Sonenshein
Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis.
J Bacteriol: 2003, 185(6);1911-22
[PubMed:12618455]
[WorldCat.org]
[DOI]
(P p)
A M Gulick, D M Schmidt, J A Gerlt, I Rayment
Evolution of enzymatic activities in the enolase superfamily: crystal structures of the L-Ala-D/L-Glu epimerases from Escherichia coli and Bacillus subtilis.
Biochemistry: 2001, 40(51);15716-24
[PubMed:11747448]
[WorldCat.org]
[DOI]
(P p)
D M Schmidt, B K Hubbard, J A Gerlt
Evolution of enzymatic activities in the enolase superfamily: functional assignment of unknown proteins in Bacillus subtilis and Escherichia coli as L-Ala-D/L-Glu epimerases.
Biochemistry: 2001, 40(51);15707-15
[PubMed:11747447]
[WorldCat.org]
[DOI]
(P p)