AbrB
- Description: transcriptional regulator of transition state genes
| Gene name | abrB |
| Synonyms | cpsX, tolB |
| Essential | no |
| Product | transcriptional regulator |
| Function | regulation of gene expression during the transition from growth to stationary phase |
| Gene expression levels in SubtiExpress: abrB | |
| Interactions involving this protein in SubtInteract: AbrB | |
| Metabolic function and regulation of this protein in SubtiPathways: Biofilm, His, Asp, Asn, Phosphorelay, Ammonium/ glutamate, Central C-metabolism, Sugar catabolism, Alternative nitrogen sources, Protein secretion | |
| MW, pI | 10 kDa, 6.57 |
| Gene length, protein length | 282 bp, 94 aa |
| Immediate neighbours | yabC, metS |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 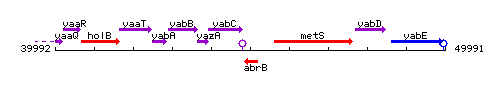
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed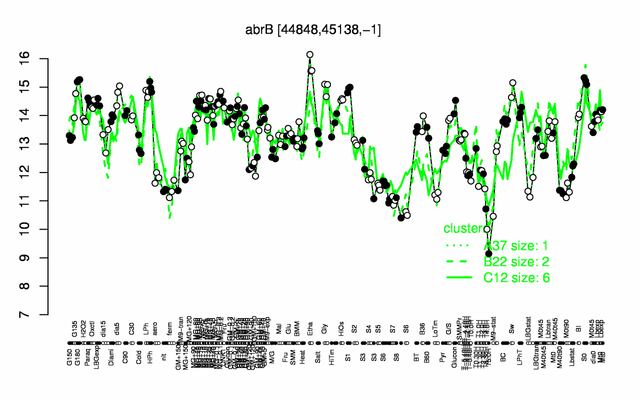
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The AbrB regulon
The gene
Basic information
- Locus tag: BSU00370
Phenotypes of a mutant
No swarming motility on B medium. PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylated on Ser-86 PubMed
- Cofactor(s):
Database entries
- UniProt: P08874
- KEGG entry: [3]
Additional information
Expression and regulation
- Operon: abrB PubMed
- Regulation: expressed at the onset of stationary phase PubMed
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Richard Losick, Harvard Univ., Cambridge, USA homepage
Mark Strauch, Baltimore, USA homepage
Your additional remarks
References
Reviews
The AbrB regulon:
Other original publications
Boumediene Soufi, Chanchal Kumar, Florian Gnad, Matthias Mann, Ivan Mijakovic, Boris Macek
Stable isotope labeling by amino acids in cell culture (SILAC) applied to quantitative proteomics of Bacillus subtilis.
J Proteome Res: 2010, 9(7);3638-46
[PubMed:20509597]
[WorldCat.org]
[DOI]
(I p)
Steve D Seredick, Barbara M Seredick, David Baker, George B Spiegelman
An A257V mutation in the bacillus subtilis response regulator Spo0A prevents regulated expression of promoters with low-consensus binding sites.
J Bacteriol: 2009, 191(17);5489-98
[PubMed:19581368]
[WorldCat.org]
[DOI]
(I p)
Yun Luo, John D Helmann
Extracytoplasmic function sigma factors with overlapping promoter specificity regulate sublancin production in Bacillus subtilis.
J Bacteriol: 2009, 191(15);4951-8
[PubMed:19465659]
[WorldCat.org]
[DOI]
(I p)
Kassem Hamze, Daria Julkowska, Sabine Autret, Krzysztof Hinc, Krzysztofa Nagorska, Agnieszka Sekowska, I Barry Holland, Simone J Séror
Identification of genes required for different stages of dendritic swarming in Bacillus subtilis, with a novel role for phrC.
Microbiology (Reading): 2009, 155(Pt 2);398-412
[PubMed:19202088]
[WorldCat.org]
[DOI]
(P p)
Frances Chu, Daniel B Kearns, Anna McLoon, Yunrong Chai, Roberto Kolter, Richard Losick
A novel regulatory protein governing biofilm formation in Bacillus subtilis.
Mol Microbiol: 2008, 68(5);1117-27
[PubMed:18430133]
[WorldCat.org]
[DOI]
(I p)
Krzysztofa Nagórska, Krzysztof Hinc, Mark A Strauch, Michał Obuchowski
Influence of the sigmaB stress factor and yxaB, the gene for a putative exopolysaccharide synthase under sigmaB Control, on biofilm formation.
J Bacteriol: 2008, 190(10);3546-56
[PubMed:18326573]
[WorldCat.org]
[DOI]
(I p)
Mark A Strauch, Benjamin G Bobay, John Cavanagh, Fude Yao, Angelo Wilson, Yoann Le Breton
Abh and AbrB control of Bacillus subtilis antimicrobial gene expression.
J Bacteriol: 2007, 189(21);7720-32
[PubMed:17720793]
[WorldCat.org]
[DOI]
(P p)
Sina Jordan, Eva Rietkötter, Mark A Strauch, Falk Kalamorz, Bronwyn G Butcher, John D Helmann, Thorsten Mascher
LiaRS-dependent gene expression is embedded in transition state regulation in Bacillus subtilis.
Microbiology (Reading): 2007, 153(Pt 8);2530-2540
[PubMed:17660417]
[WorldCat.org]
[DOI]
(P p)
Benjamin G Bobay, Geoffrey A Mueller, Richele J Thompson, Alexey G Murzin, Ronald A Venters, Mark A Strauch, John Cavanagh
NMR structure of AbhN and comparison with AbrBN: FIRST insights into the DNA binding promiscuity and specificity of AbrB-like transition state regulator proteins.
J Biol Chem: 2006, 281(30);21399-21409
[PubMed:16702211]
[WorldCat.org]
[DOI]
(P p)
Benjamin G Bobay, Antonina Andreeva, Geoffrey A Mueller, John Cavanagh, Alexey G Murzin
Revised structure of the AbrB N-terminal domain unifies a diverse superfamily of putative DNA-binding proteins.
FEBS Lett: 2005, 579(25);5669-74
[PubMed:16223496]
[WorldCat.org]
[DOI]
(P p)
Fude Yao, Mark A Strauch
Independent and interchangeable multimerization domains of the AbrB, Abh, and SpoVT global regulatory proteins.
J Bacteriol: 2005, 187(18);6354-62
[PubMed:16159768]
[WorldCat.org]
[DOI]
(P p)
Masaya Fujita, José Eduardo González-Pastor, Richard Losick
High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis.
J Bacteriol: 2005, 187(4);1357-68
[PubMed:15687200]
[WorldCat.org]
[DOI]
(P p)
Benjamin G Bobay, Linda Benson, Stephen Naylor, Brett Feeney, A Clay Clark, Michael B Goshe, Mark A Strauch, Richele Thompson, John Cavanagh
Evaluation of the DNA binding tendencies of the transition state regulator AbrB.
Biochemistry: 2004, 43(51);16106-18
[PubMed:15610005]
[WorldCat.org]
[DOI]
(P p)
Mélanie A Hamon, Nicola R Stanley, Robert A Britton, Alan D Grossman, Beth A Lazazzera
Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis.
Mol Microbiol: 2004, 52(3);847-60
[PubMed:15101989]
[WorldCat.org]
[DOI]
(P p)
Hyun-Jin Kim, Sam-In Kim, Manoja Ratnayake-Lecamwasam, Kiyoshi Tachikawa, Abraham L Sonenshein, Mark Strauch
Complex regulation of the Bacillus subtilis aconitase gene.
J Bacteriol: 2003, 185(5);1672-80
[PubMed:12591885]
[WorldCat.org]
[DOI]
(P p)
Leendert W Hamoen, Daisy Kausche, Mohamed A Marahiel, Douwe van Sinderen, Gerard Venema, Pascale Serror
The Bacillus subtilis transition state regulator AbrB binds to the -35 promoter region of comK.
FEMS Microbiol Lett: 2003, 218(2);299-304
[PubMed:12586407]
[WorldCat.org]
[DOI]
(P p)
Linda M Benson, Jeffrey L Vaughn, Mark A Strauch, Benjamin G Bobay, Richele Thompson, Stephen Naylor, John Cavanagh
Macromolecular assembly of the transition state regulator AbrB in its unbound and complexed states probed by microelectrospray ionization mass spectrometry.
Anal Biochem: 2002, 306(2);222-7
[PubMed:12123659]
[WorldCat.org]
[DOI]
(P p)
Qiang Qian, Chien Y Lee, John D Helmann, Mark A Strauch
AbrB is a regulator of the sigma(W) regulon in Bacillus subtilis.
FEMS Microbiol Lett: 2002, 211(2);219-23
[PubMed:12076816]
[WorldCat.org]
[DOI]
(P p)
Sasha H Shafikhani, Ines Mandic-Mulec, Mark A Strauch, Issar Smith, Terrance Leighton
Postexponential regulation of sin operon expression in Bacillus subtilis.
J Bacteriol: 2002, 184(2);564-71
[PubMed:11751836]
[WorldCat.org]
[DOI]
(P p)
Z E Phillips, M A Strauch
Role of Cys54 in AbrB multimerization and DNA-binding activity.
FEMS Microbiol Lett: 2001, 203(2);207-10
[PubMed:11583849]
[WorldCat.org]
[DOI]
(P p)
K Xu, M A Strauch
DNA-binding activity of amino-terminal domains of the Bacillus subtilis AbrB protein.
J Bacteriol: 2001, 183(13);4094-8
[PubMed:11395475]
[WorldCat.org]
[DOI]
(P p)
M C Gustafsson, C von Wachenfeldt
A novel diffusible substance can overcome the apparent AbrB repression of the Bacillus subtilis fatR promoter.
FEMS Microbiol Lett: 2001, 199(2);197-202
[PubMed:11377867]
[WorldCat.org]
[DOI]
(P p)
J L Vaughn, V Feher, S Naylor, M A Strauch, J Cavanagh
Novel DNA binding domain and genetic regulation model of Bacillus subtilis transition state regulator abrB.
Nat Struct Biol: 2000, 7(12);1139-46
[PubMed:11101897]
[WorldCat.org]
[DOI]
(P p)
J Hahn, A Luttinger, D Dubnau
Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis.
Mol Microbiol: 1996, 21(4);763-75
[PubMed:8878039]
[WorldCat.org]
[DOI]
(P p)
K Xu, D Clark, M A Strauch
Analysis of abrB mutations, mutant proteins, and why abrB does not utilize a perfect consensus in the -35 region of its sigma A promoter.
J Biol Chem: 1996, 271(5);2621-6
[PubMed:8576231]
[WorldCat.org]
[DOI]
(P p)
K Xu, M A Strauch
In vitro selection of optimal AbrB-binding sites: comparison to known in vivo sites indicates flexibility in AbrB binding and recognition of three-dimensional DNA structures.
Mol Microbiol: 1996, 19(1);145-58
[PubMed:8821944]
[WorldCat.org]
[DOI]
(P p)
M A Strauch
AbrB modulates expression and catabolite repression of a Bacillus subtilis ribose transport operon.
J Bacteriol: 1995, 177(23);6727-31
[PubMed:7592460]
[WorldCat.org]
[DOI]
(P p)
J Hahn, M Roggiani, D Dubnau
The major role of Spo0A in genetic competence is to downregulate abrB, an essential competence gene.
J Bacteriol: 1995, 177(12);3601-5
[PubMed:7768874]
[WorldCat.org]
[DOI]
(P p)
R Fürbass, M A Marahiel
Mutant analysis of interaction of the Bacillus subtilis transcription regulator AbrB with the antibiotic biosynthesis gene tycA.
FEBS Lett: 1991, 287(1-2);153-6
[PubMed:1908787]
[WorldCat.org]
[DOI]
(P p)
F J Slack, J P Mueller, M A Strauch, C Mathiopoulos, A L Sonenshein
Transcriptional regulation of a Bacillus subtilis dipeptide transport operon.
Mol Microbiol: 1991, 5(8);1915-25
[PubMed:1766371]
[WorldCat.org]
[DOI]
(P p)
R Fürbass, M Gocht, P Zuber, M A Marahiel
Interaction of AbrB, a transcriptional regulator from Bacillus subtilis with the promoters of the transition state-activated genes tycA and spoVG.
Mol Gen Genet: 1991, 225(3);347-54
[PubMed:1850083]
[WorldCat.org]
[DOI]
(P p)
M Strauch, V Webb, G Spiegelman, J A Hoch
The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene.
Proc Natl Acad Sci U S A: 1990, 87(5);1801-5
[PubMed:2106683]
[WorldCat.org]
[DOI]
(P p)
J B Robertson, M Gocht, M A Marahiel, P Zuber
AbrB, a regulator of gene expression in Bacillus, interacts with the transcription initiation regions of a sporulation gene and an antibiotic biosynthesis gene.
Proc Natl Acad Sci U S A: 1989, 86(21);8457-61
[PubMed:2554317]
[WorldCat.org]
[DOI]
(P p)
M A Strauch, M Perego, D Burbulys, J A Hoch
The transition state transcription regulator AbrB of Bacillus subtilis is autoregulated during vegetative growth.
Mol Microbiol: 1989, 3(9);1203-9
[PubMed:2507867]
[WorldCat.org]
[DOI]
(P p)
M A Strauch, G B Spiegelman, M Perego, W C Johnson, D Burbulys, J A Hoch
The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein.
EMBO J: 1989, 8(5);1615-21
[PubMed:2504584]
[WorldCat.org]
[DOI]
(P p)
M Perego, G B Spiegelman, J A Hoch
Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis.
Mol Microbiol: 1988, 2(6);689-99
[PubMed:3145384]
[WorldCat.org]
[DOI]
(P p)
P Zuber, R Losick
Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis.
J Bacteriol: 1987, 169(5);2223-30
[PubMed:2437099]
[WorldCat.org]
[DOI]
(P p)