LicT
- Description: transcriptional antiterminator of the bglP-bglH-yxiE operon and the bglS gene
| Gene name | licT |
| Synonyms | |
| Essential | no |
| Product | transcriptional antiterminator (BglG family) |
| Function | control of beta-glucan and beta-glucoside utilization |
| Gene expression levels in SubtiExpress: licT | |
| Interactions involving this protein in SubtInteract: LicT | |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 32 kDa, 5.944 |
| Gene length, protein length | 831 bp, 277 aa |
| Immediate neighbours | bglS, yxiP |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 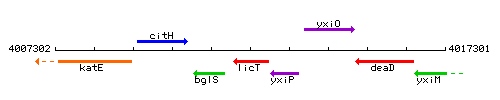
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources, transcription factors and their control, RNA binding regulators, phosphoproteins
This gene is a member of the following regulons
The LicT regulon: bglP-bglH-yxiE, bglS
The gene
Basic information
- Locus tag: BSU39080
Phenotypes of a mutant
no expression of the bglP-bglH operon
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: binding to the mRNAs of bglS and the bglP-bglH operon, causes transcription antitermination (in presence of salicin and absence of glucose)
- Protein family: transcriptional antiterminator BglG family of antiterminators (according to Swiss-Prot)
Extended information on the protein
- Kinetic information:
- K(D) for the RAT-RNA: 10 nM PubMed
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cytoplasm, even distribution in the absence of the inducer salicin, subpolar localization in the presence of salicin PubMed
Database entries
- UniProt: P39805
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP427 (licTS, erm), available in the Stülke lab
- Expression vector:
- for expression, purification of both PRDs in E. coli with N-terminal His-tag, in pWH844: pGP165, available in Stülke lab
- for expression, purification of the RNA-binding domain in E. coli with N-terminal His-tag, in pWH844: pGP315, available in Stülke lab
- for expression, purification of the RNA-binding domain in E. coli with N-terminal His-tag and thrombin cleavage site, in pGP570: pGP572, available in Stülke lab
- lacZ fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Stephane Aymerich, Microbiology and Molecular Genetics, INRA Paris-Grignon, France
Josef Deutscher, Microbiology and Molecular Genetics, INRA Paris-Grignon, France
Michael Hecker, Greifswald, Germany Homepage
Your additional remarks
References
Additional publications: PubMed
Original description
K Schnetz, J Stülke, S Gertz, S Krüger, M Krieg, M Hecker, B Rak
LicT, a Bacillus subtilis transcriptional antiterminator protein of the BglG family.
J Bacteriol: 1996, 178(7);1971-9
[PubMed:8606172]
[WorldCat.org]
[DOI]
(P p)
Control of LicT activity
Fabian M Rothe, Christoph Wrede, Martin Lehnik-Habrink, Boris Görke, Jörg Stülke Dynamic localization of a transcription factor in Bacillus subtilis: The LicT antiterminator relocalizes in response to inducer availability. J Bacteriol.: 2013, 195, Epub ahead of print. PubMed:23475962
Structural analysis of LicT
LicT-RNA interaction