YqhY
- Description: unknown
| Gene name | yqhY |
| Synonyms | |
| Essential | no PubMed |
| Product | unknown |
| Function | unknown |
| Gene expression levels in SubtiExpress: yqhY | |
| Metabolic function and regulation of this protein in SubtiPathways: yqhY | |
| MW, pI | 14 kDa, 4.414 |
| Gene length, protein length | 405 bp, 135 aa |
| Immediate neighbours | nusB, accC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 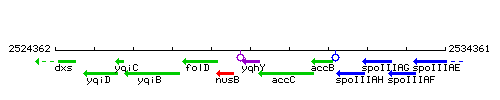
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU24330
Phenotypes of a mutant
Database entries
- BsubCyc: BSU24330
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: asp23 family (according to Swiss-Prot)
- Paralogous protein(s): YloU
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU24330
- Structure:
- UniProt: P54519
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium): 127 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 893 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 4905 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 3644 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 4928 PubMed
Biological materials
- Mutant:
- GP1468 (yqhY::erm), available in Jörg Stülke's lab PubMed
- GP1765 (yqhY::cat), available in Jörg Stülke's lab PubMed
- GP1474 (chromosomal yqhY-Strep fusion, aphA3), purification from B. subtilis, for SPINE, available in Jörg Stülke's lab
- Expression vector:
- pGP1322 (N-terminal Strep-tag, purification from E. coli, in pGP172), available in Jörg Stülke's lab
- pGP1325 (N-terminal His-tag, purification from E. coli, in pWH844), available in Jörg Stülke's lab
- pGP1496 (His-tag, purification from E. coli, in pET28a+), available in Jörg Stülke's lab
- pGP1498 (N-terminal His-tag, TEV-site, purification from E. coli, in pWH844), available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- GP1471 (spc, based on pBP43), available in Jörg Stülke's lab
- YFP fusion:
- GP1492 (cat, available in Jörg Stülke's lab
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- FLAG-tag construct:
- GP1481 (spc, based on pGP1331), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Raphael H Michna, Fabian M Commichau, Dominik Tödter, Christopher P Zschiedrich, Jörg Stülke
SubtiWiki-a database for the model organism Bacillus subtilis that links pathway, interaction and expression information.
Nucleic Acids Res: 2014, 42(Database issue);D692-8
[PubMed:24178028]
[WorldCat.org]
[DOI]
(I p)
Fabian M Commichau, Nico Pietack, Jörg Stülke
Essential genes in Bacillus subtilis: a re-evaluation after ten years.
Mol Biosyst: 2013, 9(6);1068-75
[PubMed:23420519]
[WorldCat.org]
[DOI]
(I p)
Helena B Thomaides, Ella J Davison, Lisa Burston, Hazel Johnson, David R Brown, Alison C Hunt, Jeffery Errington, Lloyd Czaplewski
Essential bacterial functions encoded by gene pairs.
J Bacteriol: 2007, 189(2);591-602
[PubMed:17114254]
[WorldCat.org]
[DOI]
(P p)
P Marini, S J Li, D Gardiol, J E Cronan, D de Mendoza
The genes encoding the biotin carboxyl carrier protein and biotin carboxylase subunits of Bacillus subtilis acetyl coenzyme A carboxylase, the first enzyme of fatty acid synthesis.
J Bacteriol: 1995, 177(23);7003-6
[PubMed:7592499]
[WorldCat.org]
[DOI]
(P p)