HtrC
- Description: similar to quality control membrane serine protease HtrA
| Gene name | htrC |
| Synonyms | yycK, yyxA |
| Essential | no |
| Product | putative quality control membrane protease |
| Function | unknown |
| Gene expression levels in SubtiExpress: htrC | |
| MW, pI | 42 kDa, 5.315 |
| Gene length, protein length | 1200 bp, 400 aa |
| Immediate neighbours | yyzO, walJ |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 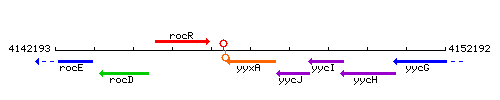
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed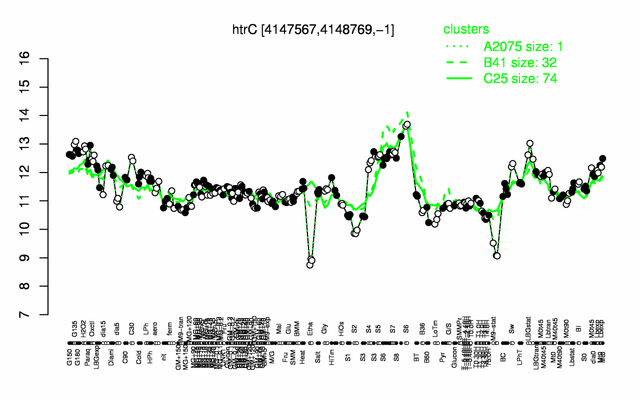
| |
Contents
Categories containing this gene/protein
proteolysis, sporulation proteins, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU40360
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: peptidase S1B family (according to Swiss-Prot)
- Paralogous protein(s): HtrA
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Interactions:
- HtrC is a member of a suspected group of hubs proteins that were suggested to be involved in a large number of interactions PubMed
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P39668
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor: for htrC: SigG PubMed
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Tim Clausen, Markus Kaiser, Robert Huber, Michael Ehrmann
HTRA proteases: regulated proteolysis in protein quality control.
Nat Rev Mol Cell Biol: 2011, 12(3);152-62
[PubMed:21326199]
[WorldCat.org]
[DOI]
(I p)
Original publications
Laxmi Krishnappa, Carmine G Monteferrante, Jolanda Neef, Annette Dreisbach, Jan Maarten van Dijl
Degradation of extracytoplasmic catalysts for protein folding in Bacillus subtilis.
Appl Environ Microbiol: 2014, 80(4);1463-8
[PubMed:24362423]
[WorldCat.org]
[DOI]
(I p)
Susanne Pohl, Gaurav Bhavsar, Joanne Hulme, Alexandra E Bloor, Goksel Misirli, Matthew W Leckenby, David S Radford, Wendy Smith, Anil Wipat, E Diane Williamson, Colin R Harwood, Rocky M Cranenburgh
Proteomic analysis of Bacillus subtilis strains engineered for improved production of heterologous proteins.
Proteomics: 2013, 13(22);3298-308
[PubMed:24115457]
[WorldCat.org]
[DOI]
(I p)
David Noone, Eric Botella, Clodagh Butler, Annette Hansen, Inga Jende, Kevin M Devine
Signal perception by the secretion stress-responsive CssRS two-component system in Bacillus subtilis.
J Bacteriol: 2012, 194(7);1800-14
[PubMed:22307758]
[WorldCat.org]
[DOI]
(I p)
Tina Wecke, Tobias Bauer, Henning Harth, Ulrike Mäder, Thorsten Mascher
The rhamnolipid stress response of Bacillus subtilis.
FEMS Microbiol Lett: 2011, 323(2);113-23
[PubMed:22092710]
[WorldCat.org]
[DOI]
(I p)
Elodie Marchadier, Rut Carballido-López, Sophie Brinster, Céline Fabret, Peggy Mervelet, Philippe Bessières, Marie-Françoise Noirot-Gros, Vincent Fromion, Philippe Noirot
An expanded protein-protein interaction network in Bacillus subtilis reveals a group of hubs: Exploration by an integrative approach.
Proteomics: 2011, 11(15);2981-91
[PubMed:21630458]
[WorldCat.org]
[DOI]
(I p)
Hein Trip, Patricia J van der Veek, Ton C Renniers, Rob Meima, Cees M Sagt, Lisette Mohrmann, Oscar P Kuipers
A novel screening system for secretion of heterologous proteins in Bacillus subtilis.
Microb Biotechnol: 2011, 4(5);673-82
[PubMed:21624103]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Hanne-Leena Hyyryläinen, Milla Pietiäinen, Tuula Lundén, Anna Ekman, Marika Gardemeister, Sanna Murtomäki-Repo, Haike Antelmann, Michael Hecker, Leena Valmu, Matti Sarvas, Vesa P Kontinen
The density of negative charge in the cell wall influences two-component signal transduction in Bacillus subtilis.
Microbiology (Reading): 2007, 153(Pt 7);2126-2136
[PubMed:17600057]
[WorldCat.org]
[DOI]
(P p)
Elise Darmon, David Noone, Anne Masson, Sierd Bron, Oscar P Kuipers, Kevin M Devine, Jan Maarten van Dijl
A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis.
J Bacteriol: 2002, 184(20);5661-71
[PubMed:12270824]
[WorldCat.org]
[DOI]
(P p)
H L Hyyryläinen, A Bolhuis, E Darmon, L Muukkonen, P Koski, M Vitikainen, M Sarvas, Z Prágai, S Bron, J M van Dijl, V P Kontinen
A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress.
Mol Microbiol: 2001, 41(5);1159-72
[PubMed:11555295]
[WorldCat.org]
[DOI]
(P p)
D Noone, A Howell, K M Devine
Expression of ykdA, encoding a Bacillus subtilis homologue of HtrA, is heat shock inducible and negatively autoregulated.
J Bacteriol: 2000, 182(6);1592-9
[PubMed:10692364]
[WorldCat.org]
[DOI]
(P p)
C Fabret, J A Hoch
A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy.
J Bacteriol: 1998, 180(23);6375-83
[PubMed:9829949]
[WorldCat.org]
[DOI]
(P p)