AraR
- Description: transcriptional repressor (GntR family, LacI family) of the arabinose utilization genes
| Gene name | araR |
| Synonyms | araC, yvbS |
| Essential | no |
| Product | transcriptional repressor (GntR family, LacI family) |
| Function | regulation of arabinose utilization
(araABDLMNPQ-abfA) and of the araE/araR genes |
| Gene expression levels in SubtiExpress: araR
(araABDLMNPQ-abfA) and of the araE/araR genes | |
| Metabolic function and regulation of this protein in SubtiPathways: AraR | |
| MW, pI | 43 kDa, 6.341 |
| Gene length, protein length | 1152 bp, 384 aa |
| Immediate neighbours | araE, yvbT |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 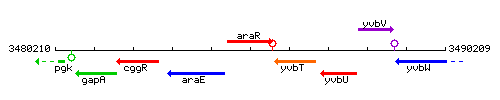
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed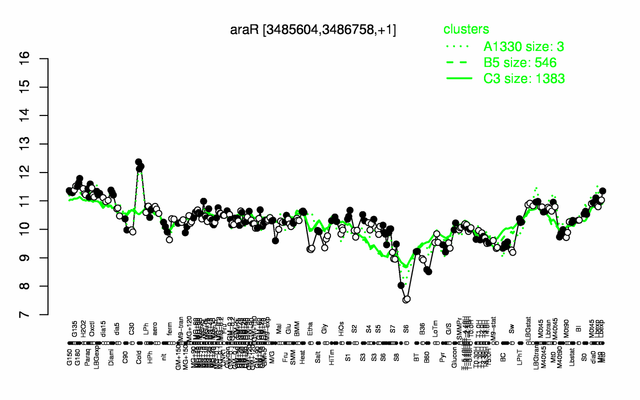
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources, transcription factors and their control
This gene is a member of the following regulons
The AraR regulon
The gene
Basic information
- Locus tag: BSU33970
Phenotypes of a mutant
Database entries
- BsubCyc: BSU33970
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: GntR family: N-terminal DNA-binding domain, LacI family: C-terminal effector-binding domain
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- N-terminal DNA-binding domain (GntR family)
- C-terminal effector-binding domain (LacI family)
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU33970
- UniProt: P96711
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Deepti Jain, Deepak T Nair
Spacing between core recognition motifs determines relative orientation of AraR monomers on bipartite operators.
Nucleic Acids Res: 2013, 41(1);639-47
[PubMed:23109551]
[WorldCat.org]
[DOI]
(I p)
Irina Saraiva Franco, Luís Jaime Mota, Cláudio Manuel Soares, Isabel de Sá-Nogueira
Probing key DNA contacts in AraR-mediated transcriptional repression of the Bacillus subtilis arabinose regulon.
Nucleic Acids Res: 2007, 35(14);4755-66
[PubMed:17617643]
[WorldCat.org]
[DOI]
(I p)
Irina Saraiva Franco, Luís Jaime Mota, Cláudio Manuel Soares, Isabel de Sá-Nogueira
Functional domains of the Bacillus subtilis transcription factor AraR and identification of amino acids important for nucleoprotein complex assembly and effector binding.
J Bacteriol: 2006, 188(8);3024-36
[PubMed:16585763]
[WorldCat.org]
[DOI]
(P p)
Maria Paiva Raposo, José Manuel Inácio, Luís Jaime Mota, Isabel de Sá-Nogueira
Transcriptional regulation of genes encoding arabinan-degrading enzymes in Bacillus subtilis.
J Bacteriol: 2004, 186(5);1287-96
[PubMed:14973026]
[WorldCat.org]
[DOI]
(P p)
L J Mota, L M Sarmento, I de Sá-Nogueira
Control of the arabinose regulon in Bacillus subtilis by AraR in vivo: crucial roles of operators, cooperativity, and DNA looping.
J Bacteriol: 2001, 183(14);4190-201
[PubMed:11418559]
[WorldCat.org]
[DOI]
(P p)
L J Mota, P Tavares, I Sá-Nogueira
Mode of action of AraR, the key regulator of L-arabinose metabolism in Bacillus subtilis.
Mol Microbiol: 1999, 33(3);476-89
[PubMed:10417639]
[WorldCat.org]
[DOI]
(P p)
I Sá-Nogueira, L J Mota
Negative regulation of L-arabinose metabolism in Bacillus subtilis: characterization of the araR (araC) gene.
J Bacteriol: 1997, 179(5);1598-608
[PubMed:9045819]
[WorldCat.org]
[DOI]
(P p)
I Sá-Nogueira, H Paveia, H de Lencastre
Isolation of constitutive mutants for L-arabinose utilization in Bacillus subtilis.
J Bacteriol: 1988, 170(6);2855-7
[PubMed:3131313]
[WorldCat.org]
[DOI]
(P p)