McpA
- Description: membrane-bound chemotaxis receptor, methyl-accepting chemotaxis protein
| Gene name | mcpA |
| Synonyms | |
| Essential | no |
| Product | methyl-accepting chemotaxis protein |
| Function | control of chemotaxis |
| Gene expression levels in SubtiExpress: mcpA | |
| Interactions involving this protein in SubtInteract: McpA | |
| MW, pI | 72 kDa, 4.988 |
| Gene length, protein length | 1983 bp, 661 aa |
| Immediate neighbours | tlpB, tlpA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed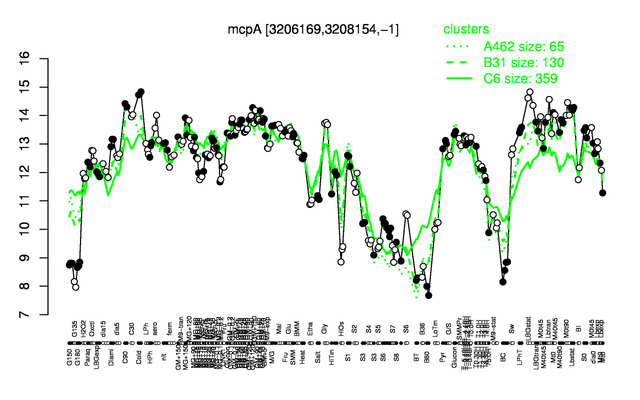
| |
Contents
Categories containing this gene/protein
motility and chemotaxis, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU31240
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): McpB
Extended information on the protein
- Kinetic information:
- Domains:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane PubMed
Database entries
- Structure:
- UniProt: P39214
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
- in minimal medium, McpA is present with 15,900 +/- 3,000 molecules per cell PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
George D Glekas, Matthew J Plutz, Hanna E Walukiewicz, George M Allen, Christopher V Rao, George W Ordal
Elucidation of the multiple roles of CheD in Bacillus subtilis chemotaxis.
Mol Microbiol: 2012, 86(3);743-56
[PubMed:22931217]
[WorldCat.org]
[DOI]
(I p)
Vincent J Cannistraro, George D Glekas, Christopher V Rao, George W Ordal
Cellular stoichiometry of the chemotaxis proteins in Bacillus subtilis.
J Bacteriol: 2011, 193(13);3220-7
[PubMed:21515776]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
D W Hanlon, G W Ordal
Cloning and characterization of genes encoding methyl-accepting chemotaxis proteins in Bacillus subtilis.
J Biol Chem: 1994, 269(19);14038-46
[PubMed:8188684]
[WorldCat.org]
(P p)
D W Hanlon, C Ying, G W Ordal
Purification and reconstitution of the methyl-accepting chemotaxis proteins from Bacillus subtilis.
Biochim Biophys Acta: 1993, 1158(3);345-51
[PubMed:8251536]
[WorldCat.org]
[DOI]
(P p)
M S Thoelke, J M Casper, G W Ordal
Methyl group turnover on methyl-accepting chemotaxis proteins during chemotaxis by Bacillus subtilis.
J Biol Chem: 1990, 265(4);1928-32
[PubMed:2105313]
[WorldCat.org]
(P p)
M S Thoelke, J R Kirby, G W Ordal
Novel methyl transfer during chemotaxis in Bacillus subtilis.
Biochemistry: 1989, 28(13);5585-9
[PubMed:2505839]
[WorldCat.org]
[DOI]
(P p)
J A Ahlgren, G W Ordal
Methyl esterification of glutamic acid residues of methyl-accepting chemotaxis proteins in Bacillus subtilis.
Biochem J: 1983, 213(3);759-63
[PubMed:6137212]
[WorldCat.org]
[DOI]
(P p)