TsaC
Revision as of 14:05, 13 May 2013 by 134.76.70.252 (talk)
- Description: L-threonylcarbamoyl-AMP synthase, biosynthesis of the hypermodified base threonylcarbamoyladenosine (t(6)A) (together with TsaB-TsaD-TsaE)
| Gene name | tsaC |
| Synonyms | ipc-29d, ywlC |
| Essential | no |
| Product | L-threonylcarbamoyl-AMP synthase |
| Function | tRNA modification |
| Gene expression levels in SubtiExpress: tsaC | |
| MW, pI | 36 kDa, 5.82 |
| Gene length, protein length | 1038 bp, 346 aa |
| Immediate neighbours | ywlD, ywlB |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 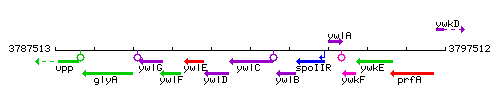
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed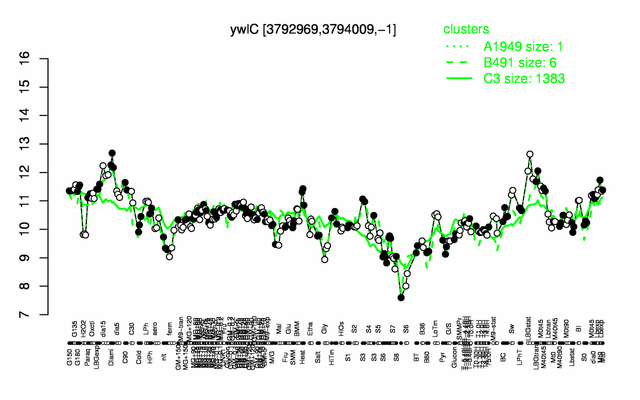
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU36950
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- conversion of L-threonine, bicarbonate/CO2 and ATP to give the intermediate L-threonylcarbamoyl-AMP (TC-AMP) and pyrophosphate as products PubMed
- biosynthesis of the hypermodified base threonylcarbamoyladenosine (t(6)A), a universal modification found at position 37 of ANN decoding tRNAs, which imparts a unique structure to the anticodon loop enhancing its binding to ribosomes in vitro (shown for the corresponding E. coli protein YrdC) PubMed
- Protein family: SUA5 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P39153
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References