TyrS
- Description: tyrosyl-tRNA synthetase (major)
| Gene name | tyrS |
| Synonyms | |
| Essential | yes PubMed |
| Product | tyrosyl-tRNA synthetase (major) |
| Function | translation |
| Gene expression levels in SubtiExpress: tyrS | |
| Metabolic function and regulation of this protein in SubtiPathways: tyrS | |
| MW, pI | 47 kDa, 5.213 |
| Gene length, protein length | 1266 bp, 422 aa |
| Immediate neighbours | rpsD, ytzK |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 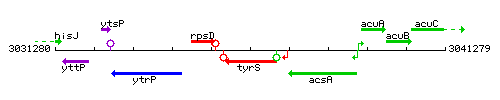
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed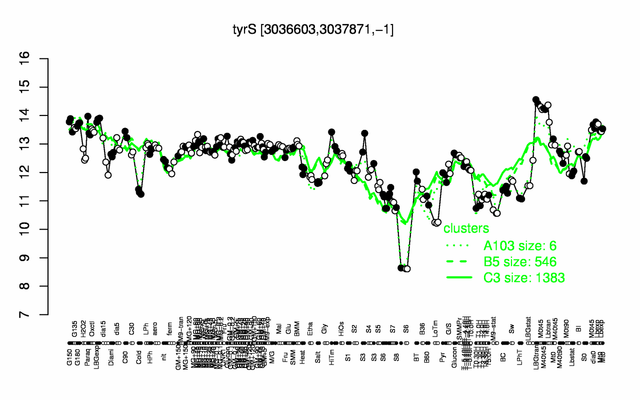
| |
Contents
Categories containing this gene/protein
translation, essential genes, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29670
Phenotypes of a mutant
- essential PubMed
- the gene can be deleted, but the mutant acquires suppressors in dtrR to allow expression of tyrZ PubMed
- a tyrS tyrZ double mutant is not viable PubMed
Database entries
- BsubCyc: BSU29670
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU29670
- Structure:
- UniProt: P22326
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- T-box: RNA switch, transcriptional antitermination PubMed
- Additional information:
- belongs to the 100 most abundant proteins PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 848 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 3406 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 4314 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 1369 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 1521 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications