Nap
- Description: carboxylesterase NA
| Gene name | nap |
| Synonyms | cesA |
| Essential | no |
| Product | carboxylesterase NA |
| Function | lipid degradation |
| Gene expression levels in SubtiExpress: nap | |
| MW, pI | 33 kDa, 5.798 |
| Gene length, protein length | 900 bp, 300 aa |
| Immediate neighbours | ydfJ, ydfK |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 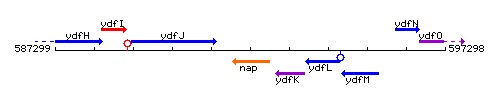
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed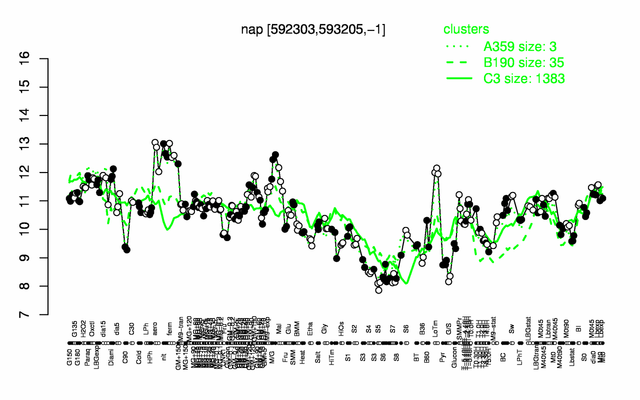
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU05440
Phenotypes of a mutant
Database entries
- BsubCyc: BSU05440
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: A carboxylic ester + H2O = an alcohol + a carboxylate (according to Swiss-Prot)
- Protein family: AB hydrolase superfamily (according to Swiss-Prot)
- Paralogous protein(s): YbfK
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU05440
- Structure: 2R11
- UniProt: P96688
- KEGG entry: [3]
- E.C. number: 3.1.1.1 3.1.1.1]
Additional information
Expression and regulation
- Operon: nap PubMed
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Henriëtte J Rozeboom, Luis F Godinho, Marco Nardini, Wim J Quax, Bauke W Dijkstra
Crystal structures of two Bacillus carboxylesterases with different enantioselectivities.
Biochim Biophys Acta: 2014, 1844(3);567-75
[PubMed:24418394]
[WorldCat.org]
[DOI]
(P p)
Luis F Godinho, Carlos R Reis, Henriëtte J Rozeboom, Frank J Dekker, Bauke W Dijkstra, Gerrit J Poelarends, Wim J Quax
Enhancement of the enantioselectivity of carboxylesterase A by structure-based mutagenesis.
J Biotechnol: 2012, 158(1-2);36-43
[PubMed:22248594]
[WorldCat.org]
[DOI]
(I p)
Ken-ichi Yoshida, Hirotake Yamaguchi, Masaki Kinehara, Yo-hei Ohki, Yoshiko Nakaura, Yasutaro Fujita
Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box.
Mol Microbiol: 2003, 49(1);157-65
[PubMed:12823818]
[WorldCat.org]
[DOI]
(P p)