SigD
- Description: RNA polymerase sigma factor SigD
| Gene name | sigD |
| Synonyms | flaB |
| Essential | no |
| Product | RNA polymerase sigma factor SigD |
| Function | regulation of flagella, motility, chemotaxis and autolysis |
| Gene expression levels in SubtiExpress: sigD | |
| Interactions involving this protein in SubtInteract: SigD | |
| MW, pI | 29 kDa, 5.167 |
| Gene length, protein length | 762 bp, 254 aa |
| Immediate neighbours | cheD, swrB |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 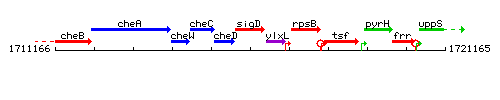
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
transcription, sigma factors and their control, motility and chemotaxis
This gene is a member of the following regulons
CodY regulon, SigD regulon, Spo0A regulon
The SigD regulon
The gene
Basic information
- Locus tag: BSU16470
Phenotypes of a mutant
- Inactivation of alsR, sigD and sigW increases competitive fitness of Bacillus subtilis under non-sporulating conditions PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: sigma-70 factor family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
Database entries
- Structure:
- UniProt: P10726
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- flgB-flgC-fliE-fliF-fliG-fliH-fliI-fliJ-ylxF-fliK-flgD-flgE-fliL-fliM-fliY-cheY-fliZ-fliP-fliQ-fliR-flhB-flhA-flhF-flhG-cheB-cheA-cheW-cheC-cheD-sigD-swrB PubMed
- ylxF-fliK-flgD-flgE-fliL-fliM-fliY-cheY-fliZ-fliP-fliQ-fliR-flhB-flhA-flhF-flhG-cheB-cheA-cheW-cheC-cheD-sigD-swrB PubMed
- sigD-swrB PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
The SigD regulon
Original publications
Additional publications: PubMed
Diethmaier C, Pietack N, Gunka K, Wrede C, Lehnik-Habrink M, Herzberg C, Hübner S, Stülke J A Novel Factor Controlling Bistability in Bacillus subtilis: The YmdB Protein Affects Flagellin Expression and Biofilm Formation. J Bacteriol.: 2011, 193(21):5997-6007. PubMed:21856853
Loralyn M Cozy, Daniel B Kearns
Gene position in a long operon governs motility development in Bacillus subtilis.
Mol Microbiol: 2010, 76(2);273-85
[PubMed:20233303]
[WorldCat.org]
[DOI]
(I p)
Letal I Salzberg, John D Helmann
Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition.
J Bacteriol: 2008, 190(23);7797-807
[PubMed:18820022]
[WorldCat.org]
[DOI]
(I p)
Kazuo Kobayashi
Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis.
Mol Microbiol: 2007, 66(2);395-409
[PubMed:17850253]
[WorldCat.org]
[DOI]
(P p)
Ana R Perez, Angelica Abanes-De Mello, Kit Pogliano
Suppression of engulfment defects in bacillus subtilis by elevated expression of the motility regulon.
J Bacteriol: 2006, 188(3);1159-64
[PubMed:16428420]
[WorldCat.org]
[DOI]
(P p)
Daniel B Kearns, Richard Losick
Cell population heterogeneity during growth of Bacillus subtilis.
Genes Dev: 2005, 19(24);3083-94
[PubMed:16357223]
[WorldCat.org]
[DOI]
(P p)
H Werhane, P Lopez, M Mendel, M Zimmer, G W Ordal, L M Márquez-Magaña
The last gene of the fla/che operon in Bacillus subtilis, ylxL, is required for maximal sigmaD function.
J Bacteriol: 2004, 186(12);4025-9
[PubMed:15175317]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
Haike Antelmann, Hiroki Yamamoto, Junichi Sekiguchi, Michael Hecker
Stabilization of cell wall proteins in Bacillus subtilis: a proteomic approach.
Proteomics: 2002, 2(5);591-602
[PubMed:11987133]
[WorldCat.org]
[DOI]
(P p)
D B Mirel, W F Estacio, M Mathieu, E Olmsted, J Ramirez, L M Márquez-Magaña
Environmental regulation of Bacillus subtilis sigma(D)-dependent gene expression.
J Bacteriol: 2000, 182(11);3055-62
[PubMed:10809682]
[WorldCat.org]
[DOI]
(P p)
M G Bertero, B Gonzales, C Tarricone, F Ceciliani, A Galizzi
Overproduction and characterization of the Bacillus subtilis anti-sigma factor FlgM.
J Biol Chem: 1999, 274(17);12103-7
[PubMed:10207036]
[WorldCat.org]
[DOI]
(P p)
W Estacio, S S Anna-Arriola, M Adedipe, L M Márquez-Magaña
Dual promoters are responsible for transcription initiation of the fla/che operon in Bacillus subtilis.
J Bacteriol: 1998, 180(14);3548-55
[PubMed:9657996]
[WorldCat.org]
[DOI]
(P p)
T Caramori, A Galizzi
The UP element of the promoter for the flagellin gene, hag, stimulates transcription from both SigD- and SigA-dependent promoters in Bacillus subtilis.
Mol Gen Genet: 1998, 258(4);385-8
[PubMed:9648743]
[WorldCat.org]
[DOI]
(P p)
R Allmansberger
Temporal regulation of sigD from Bacillus subtilis depends on a minor promoter in front of the gene.
J Bacteriol: 1997, 179(20);6531-5
[PubMed:9335309]
[WorldCat.org]
[DOI]
(P p)
K Fredrick, J D Helmann
RNA polymerase sigma factor determines start-site selection but is not required for upstream promoter element activation on heteroduplex (bubble) templates.
Proc Natl Acad Sci U S A: 1997, 94(10);4982-7
[PubMed:9144176]
[WorldCat.org]
[DOI]
(P p)
Y F Chen, J D Helmann
DNA-melting at the Bacillus subtilis flagellin promoter nucleates near -10 and expands unidirectionally.
J Mol Biol: 1997, 267(1);47-59
[PubMed:9096206]
[WorldCat.org]
[DOI]
(P p)
K Fredrick, J D Helmann
FlgM is a primary regulator of sigmaD activity, and its absence restores motility to a sinR mutant.
J Bacteriol: 1996, 178(23);7010-3
[PubMed:8955328]
[WorldCat.org]
[DOI]
(P p)
M H Rashid, J Sekiguchi
flaD (sinR) mutations affect SigD-dependent functions at multiple points in Bacillus subtilis.
J Bacteriol: 1996, 178(22);6640-3
[PubMed:8932324]
[WorldCat.org]
[DOI]
(P p)
Y F Chen, J D Helmann
The Bacillus subtilis flagellar regulatory protein sigma D: overproduction, domain analysis and DNA-binding properties.
J Mol Biol: 1995, 249(4);743-53
[PubMed:7602586]
[WorldCat.org]
[DOI]
(P p)
K Fredrick, T Caramori, Y F Chen, A Galizzi, J D Helmann
Promoter architecture in the flagellar regulon of Bacillus subtilis: high-level expression of flagellin by the sigma D RNA polymerase requires an upstream promoter element.
Proc Natl Acad Sci U S A: 1995, 92(7);2582-6
[PubMed:7708689]
[WorldCat.org]
[DOI]
(P p)
L Chen, J D Helmann
The Bacillus subtilis sigma D-dependent operon encoding the flagellar proteins FliD, FliS, and FliT.
J Bacteriol: 1994, 176(11);3093-101
[PubMed:8195064]
[WorldCat.org]
[DOI]
(P p)
K L Fredrick, J D Helmann
Dual chemotaxis signaling pathways in Bacillus subtilis: a sigma D-dependent gene encodes a novel protein with both CheW and CheY homologous domains.
J Bacteriol: 1994, 176(9);2727-35
[PubMed:8169223]
[WorldCat.org]
[DOI]
(P p)
L M Márquez-Magaña, M J Chamberlin
Characterization of the sigD transcription unit of Bacillus subtilis.
J Bacteriol: 1994, 176(8);2427-34
[PubMed:8157612]
[WorldCat.org]
[DOI]
(P p)
D B Mirel, M J Chamberlin
The Bacillus subtilis flagellin gene (hag) is transcribed by the sigma 28 form of RNA polymerase.
J Bacteriol: 1989, 171(6);3095-101
[PubMed:2498284]
[WorldCat.org]
[DOI]
(P p)
J D Helmann, F R Masiarz, M J Chamberlin
Isolation and characterization of the Bacillus subtilis sigma 28 factor.
J Bacteriol: 1988, 170(4);1560-7
[PubMed:3127378]
[WorldCat.org]
[DOI]
(P p)
J D Helmann, L M Márquez, M J Chamberlin
Cloning, sequencing, and disruption of the Bacillus subtilis sigma 28 gene.
J Bacteriol: 1988, 170(4);1568-74
[PubMed:2832368]
[WorldCat.org]
[DOI]
(P p)