PtsG
- Description: trigger enzyme: major glucose permease of the PTS, EIICBA(Glc) and control of GlcT activity
| Gene name | ptsG |
| Synonyms | ptsX, crr |
| Essential | no |
| Product | trigger enzyme: glucose-specific enzyme IICBA component of the PTS |
| Function | glucose transport and phosphorylation, control of GlcT activity |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism, Sugar catabolism | |
| MW, pI | 75,3 kDa, 5.40 |
| Gene length, protein length | 2097 bp, 699 amino acids |
| Immediate neighbours | glcT, ptsH |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 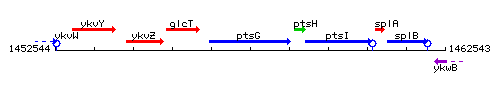
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU13890
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transport and phosphorylation of glucose, receives a phosphate from HPr at the IIA domain (His-620), the phosphate group is then transferred to the IIB domain (Cys-461) an finally to the incoming glucose. In the absence of glucose, PtsG phosphorylates and thereby inactivates the transcriptional antiterminator GlcT.
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- 11x transmembrane domain (16–36, 89–109, 139–159, 180–200, 233–253, 283–303, 313–333, 338–358, 365–385, 388–408)
- PTS EIIC domain ( 1-424)
- PTS EIIB domain (439–520)
- PTS EIIA domain (568–672)
- Modification: transient phosphorylation (HPr-dependent) on His-620, then internal phosphotransfer from His-620 to Cys-461
- Cofactor(s):
- Effectors of protein activity:
- Localization: membrane protein PubMed
Database entries
- UniProt: P20166
- KEGG entry: [3]
- E.C. number: 2.7.1.69
Additional information
Expression and regulation
- Regulatory mechanism: transcriptional antitermination via the GlcT-dependent RNA switch PubMed
- Additional information:
Biological materials
- Mutant: GP474 (cat), QB5436 (spc), QB5445 (erm), available in Stülke lab
- Expression vector:
- lacZ fusion:
- pGP34 (pAC5), available in Stülke lab
- pGP66 (pAC7), available in Stülke lab
- pGP606 (mutant terminator, pAC6), available in Stülke lab
- pGP532 (pAC7), available in Stülke lab
- series of promoter deletions are available in pAC5 and pAC6, available in Stülke lab
- series of RAT mutants are available in pAC6, available in Stülke lab
- GFP fusion:
- Antibody:
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References