Difference between revisions of "MalP"
| Line 123: | Line 123: | ||
<pubmed>,16707683,10627040 10627040, </pubmed> | <pubmed>,16707683,10627040 10627040, </pubmed> | ||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 13:42, 15 June 2009
- Description: maltose-specific phosphotransferase system, EIICB
| Gene name | malP |
| Synonyms | glvC |
| Essential | no |
| Product | maltose-specific phosphotransferase system, EIICB |
| Function | maltose uptake and phosphorylation |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 57 kDa, 8.872 |
| Gene length, protein length | 1581 bp, 527 aa |
| Immediate neighbours | glvR, yfiB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 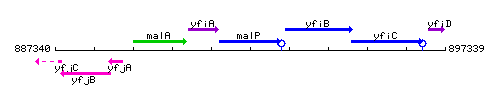
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU08200
Phenotypes of a mutant
no growth on maltose as single carbon source PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Protein EIIB N(pi)-phospho-L-histidine/cysteine + sugar = protein EIIB + sugar phosphate (according to Swiss-Prot)
- Protein family: PTS permease, glucose permease (Glc) family PubMed
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- Swiss prot entry: P54715
- KEGG entry: [2]
- E.C. number: 2.7.1.69
Additional information
Expression and regulation
- Regulation: repressed by glucose (CcpA)
- Regulatory mechanism: CcpA: transcription repression
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Stefan Schönert, Sabine Seitz, Holger Krafft, Eva-Anne Feuerbaum, Iris Andernach, Gabriele Witz, Michael K Dahl
Maltose and maltodextrin utilization by Bacillus subtilis.
J Bacteriol: 2006, 188(11);3911-22
[PubMed:16707683]
[WorldCat.org]
[DOI]
(P p)
Jonathan Reizer, Steffi Bachem, Aiala Reizer, Maryvonne Arnaud, Milton H Saier, Jörg Stülke
Novel phosphotransferase system genes revealed by genome analysis - the complete complement of PTS proteins encoded within the genome of Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 12);3419-3429
[PubMed:10627040]
[WorldCat.org]
[DOI]
(P p)