Difference between revisions of "YjmB"
| Line 88: | Line 88: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[uxaC]]-[[yjmB]]-[[yjmC]]-[[yjmD]]-[[uxuA]]-[[yjmF]]-[[exuT]]-[[exuR]]-[[uxaB]]-[[uxaA]]'' [http://www.ncbi.nlm.nih.gov/sites/entrez/9882655 PubMed] |
| − | * '''[[Sigma factor]]:''' | + | * '''[[Sigma factor]]:''' [[SigA]] |
| − | * '''Regulation:''' repressed by glucose ([[CcpA]]) | + | * '''Regulation:''' |
| + | ** repressed by glucose ([[CcpA]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/9882655 PubMed1] [http://www.ncbi.nlm.nih.gov/sites/entrez/10666464 PubMed2] | ||
| + | ** induced by galacturonate ([[ExuR]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/9882655 PubMed] | ||
| − | * '''Regulatory mechanism:''' [[CcpA]]: transcription repression | + | * '''Regulatory mechanism:''' |
| + | ** [[CcpA]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/9882655 PubMed1] [http://www.ncbi.nlm.nih.gov/sites/entrez/10666464 PubMed2] | ||
| + | ** [[ExuR]]: reanscription repression in the absence of inducer [http://www.ncbi.nlm.nih.gov/sites/entrez/9882655 PubMed] | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
| Line 118: | Line 122: | ||
=References= | =References= | ||
| − | + | <pubmed> 9579062, 9882655, 10666464 </pubmed> | |
# Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | ||
Revision as of 13:48, 30 May 2009
- Description: similar to Na+:galactoside symporter
| Gene name | yjmB |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | unknown |
| MW, pI | 50 kDa, 9.83 |
| Gene length, protein length | 1377 bp, 459 aa |
| Immediate neighbours | uxaC, yjmC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 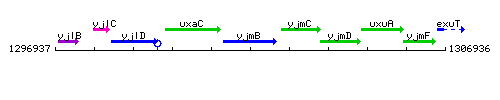
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: View classification (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- Swiss prot entry: O34961
- KEGG entry: BSU12310
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Y Miwa, A Nakata, A Ogiwara, M Yamamoto, Y Fujita
Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis.
Nucleic Acids Res: 2000, 28(5);1206-10
[PubMed:10666464]
[WorldCat.org]
[DOI]
(I p)
K R Mekjian, E M Bryan, B W Beall, C P Moran
Regulation of hexuronate utilization in Bacillus subtilis.
J Bacteriol: 1999, 181(2);426-33
[PubMed:9882655]
[WorldCat.org]
[DOI]
(P p)
Carlo Rivolta, Blazenka Soldo, Vladimir Lazarevic, Bernard Joris, Catherine Mauël, Dimitri Karamat
A 35.7 kb DNA fragment from the Bacillus subtilis chromosome containing a putative 12.3 kb operon involved in hexuronate catabolism and a perfectly symmetrical hypothetical catabolite-responsive element.
Microbiology (Reading): 1998, 144 ( Pt 4);877-884
[PubMed:9579062]
[WorldCat.org]
[DOI]
(P p)
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed