Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' glyceraldehyde-3-phosphate dehydrogenase, NADP-dependent, gluconeogenic enzyme<br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''gapB'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''ppc'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | + | |style="background:#ABCDEF;" align="center"| '''Essential''' || no |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || glyceraldehyde-3-phosphate dehydrogenase 2 |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || anabolic enzyme in gluconeogenesis |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 37,3 kDa, 6.47 |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 1020 bp, 340 amino acids |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ytcD]]'', ''[[speD]]'' |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"|'''Hier soll was neues rein''' | |colspan="2" style="background:#FAF8CC;" align="center"|'''Hier soll was neues rein''' | ||
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |colspan="2" | '''Genetic context''' <br/> [[Image:gapB_context.gif]] |
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| Line 35: | Line 35: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Coordinates:''' | + | * '''Coordinates:''' 2966075 - 2967094 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| Line 41: | Line 41: | ||
=== Database entries === | === Database entries === | ||
| − | * '''DBTBS entry:''' | + | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/gapB-speD.html] |
| − | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+ | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG12592] |
=== Additional information=== | === Additional information=== | ||
| − | |||
=The protein= | =The protein= | ||
| Line 52: | Line 51: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' D-glyceraldehyde 3-phosphate + phosphate + NAD(P)(+) = 3-phospho-D-glyceroyl phosphate + NAD(P)H. |
| − | * '''Protein family:''' | + | * '''Protein family:''' glyceraldehyde-3-phosphate dehydrogenase family |
| − | * '''Paralogous protein(s):''' | + | * '''Paralogous protein(s):''' [[GapA]] |
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 63: | Line 62: | ||
* '''Domains:''' | * '''Domains:''' | ||
| + | ** Nucleotid bindinge domain (12-13) | ||
| + | ** 2x Glyceraldehyde 3-phosphate binding domain (151-153) & (210-211) | ||
* '''Modification:''' | * '''Modification:''' | ||
| Line 72: | Line 73: | ||
* '''Interactions:''' | * '''Interactions:''' | ||
| − | * '''Localization:''' | + | * '''Localization:''' Cytoplasm (Homogeneous) [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] |
=== Database entries === | === Database entries === | ||
| Line 78: | Line 79: | ||
* '''Structure:''' | * '''Structure:''' | ||
| − | * '''Swiss prot entry:''' | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/O34425] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU29020] |
| − | * '''E.C. number:''' | + | * '''E.C. number:''' [http://www.expasy.ch/cgi-bin/get-enzyme-entry?1.2.1.12 1.2.1.12] |
=== Additional information=== | === Additional information=== | ||
| + | |||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[gapB]]-[[speD]]'' |
| − | * ''' | + | * '''Sigma factor:''' [[SigA]] [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+15720552 PubMed] |
| − | * '''Regulation:''' | + | * '''Regulation:''' repressed (70-times) by Glc, repressor [[CcpN]] [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+15720552 PubMed] |
| − | * '''Regulatory mechanism:''' | + | * '''Regulatory mechanism:''' transcription repression |
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
| Line 103: | Line 105: | ||
* '''Expression vector:''' | * '''Expression vector:''' | ||
| − | + | ||
* '''lacZ fusion:''' | * '''lacZ fusion:''' | ||
* '''GFP fusion:''' | * '''GFP fusion:''' | ||
| − | |||
| − | |||
* '''Antibody:''' | * '''Antibody:''' | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| + | |||
| + | [[Stephane Aymerich |Stephane Aymerich]], Microbiology and Molecular Genetics, INRA Paris-Grignon, France | ||
=Your additional remarks= | =Your additional remarks= | ||
| Line 118: | Line 120: | ||
=References= | =References= | ||
| − | # | + | # Fillinger, S., Boschi-Muller, S., Azza, S., Dervyn, E., Branlant, G., and Aymerich, S. (2000) Two glyceraldehyde-3-phosphate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium. J Biol Chem 275, 14031-14037. [http://www.ncbi.nlm.nih.gov/sites/entrez/10799476 PubMed] |
| + | # Meile et al. (2006) Systematic localisation of proteins fused to the green fluorescent protein in ''Bacillus subtilis'': identification of new proteins at the DNA replication factory ''Proteomics'' '''6:''' 2135-2146. [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] | ||
| + | # Servant et al. (2005) CcpN (YqzB), a novel regulator for CcpA-independent catabolite repression of ''Bacillus subtilis'' gluconeogenic genes. Mol. Microbiol. 55: 1435-1451. [http://www.ncbi.nlm.nih.gov/sites/entrez/15720552 PubMed] | ||
| + | # Tännler et al. (2008) CcpN controls central carbon fluxes in ''Bacillus subtilis''. J. Bacteriol. 190: 6178-6187. [http://www.ncbi.nlm.nih.gov/sites/entrez/18586936 PubMed] | ||
| + | # Thomaides, H. B., Davison, E. J., Burston, L., Johnson, H., Brown, D. R., Hunt, A. C., Errington, J., and Czaplewski, L. (2007) Essential bacterial functions encoded by gene pairs. J Bacteriol 189: 591-602. [http://www.ncbi.nlm.nih.gov/sites/entrez/17114254 PubMed] | ||
Revision as of 17:19, 15 April 2009
- Description: glyceraldehyde-3-phosphate dehydrogenase, NADP-dependent, gluconeogenic enzyme
| Gene name | gapB |
| Synonyms | ppc |
| Essential | no |
| Product | glyceraldehyde-3-phosphate dehydrogenase 2 |
| Function | anabolic enzyme in gluconeogenesis |
| MW, pI | 37,3 kDa, 6.47 |
| Gene length, protein length | 1020 bp, 340 amino acids |
| Immediate neighbours | ytcD, speD |
| Hier soll was neues rein | |
Genetic context 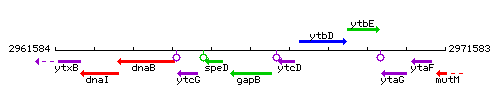
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates: 2966075 - 2967094
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: D-glyceraldehyde 3-phosphate + phosphate + NAD(P)(+) = 3-phospho-D-glyceroyl phosphate + NAD(P)H.
- Protein family: glyceraldehyde-3-phosphate dehydrogenase family
- Paralogous protein(s): GapA
Extended information on the protein
- Kinetic information:
- Domains:
- Nucleotid bindinge domain (12-13)
- 2x Glyceraldehyde 3-phosphate binding domain (151-153) & (210-211)
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: Cytoplasm (Homogeneous) PubMed
Database entries
- Structure:
- Swiss prot entry: [3]
- KEGG entry: [4]
- E.C. number: 1.2.1.12
Additional information
Expression and regulation
- Regulatory mechanism: transcription repression
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- Antibody:
Labs working on this gene/protein
Stephane Aymerich, Microbiology and Molecular Genetics, INRA Paris-Grignon, France
Your additional remarks
References
- Fillinger, S., Boschi-Muller, S., Azza, S., Dervyn, E., Branlant, G., and Aymerich, S. (2000) Two glyceraldehyde-3-phosphate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium. J Biol Chem 275, 14031-14037. PubMed
- Meile et al. (2006) Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory Proteomics 6: 2135-2146. PubMed
- Servant et al. (2005) CcpN (YqzB), a novel regulator for CcpA-independent catabolite repression of Bacillus subtilis gluconeogenic genes. Mol. Microbiol. 55: 1435-1451. PubMed
- Tännler et al. (2008) CcpN controls central carbon fluxes in Bacillus subtilis. J. Bacteriol. 190: 6178-6187. PubMed
- Thomaides, H. B., Davison, E. J., Burston, L., Johnson, H., Brown, D. R., Hunt, A. C., Errington, J., and Czaplewski, L. (2007) Essential bacterial functions encoded by gene pairs. J Bacteriol 189: 591-602. PubMed