Difference between revisions of "YfhM"
| Line 92: | Line 92: | ||
* '''Operon:''' | * '''Operon:''' | ||
| − | * '''Sigma factor:''' [[SigB]] [http://www.ncbi.nlm.nih.gov/pubmed/15805528 PubMed] | + | * '''[[Sigma factor]]:''' [[SigB]] [http://www.ncbi.nlm.nih.gov/pubmed/15805528 PubMed] |
* '''Regulation:''' induced by stress ([[SigB]]) [http://www.ncbi.nlm.nih.gov/pubmed/15805528 PubMed] | * '''Regulation:''' induced by stress ([[SigB]]) [http://www.ncbi.nlm.nih.gov/pubmed/15805528 PubMed] | ||
Revision as of 10:01, 8 April 2009
- Description: general stress protein, similar to epoxide hydrolase
| Gene name | yfhM |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | survival of ethanol stress |
| MW, pI | 32 kDa, 6.064 |
| Gene length, protein length | 858 bp, 286 aa |
| Immediate neighbours | yfhL, csbB |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 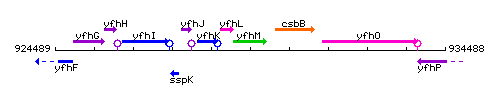
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- Swiss prot entry:
- KEGG entry: [3]
- E.C. number:
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Höper et al. (2005) Comprehensive Characterization of the Contribution of Individual SigB-Dependent General Stress Genes to Stress Resistance of Bacillus subtilis. J. Bact. 187: 2810-2826 PubMed
- Gerth et al. (2008) Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J Bacteriol 190:321-331 PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed