Difference between revisions of "GyrB"
| Line 1: | Line 1: | ||
| − | * '''Description:''' DNA- | + | * '''Description:''' DNA-Gyrase (subunit B) <br/><br/> |
| − | |||
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| − | |- | + | |- |
| − | |||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
|''gyrB'' | |''gyrB'' | ||
| − | |- | + | |- |
| − | |||
|style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''novA '' | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''novA '' | ||
| − | |- | + | |- |
| − | |||
|style="background:#ABCDEF;" align="center"| '''Essential''' || yes [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed] | |style="background:#ABCDEF;" align="center"| '''Essential''' || yes [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed] | ||
| − | |- | + | |- |
| − | + | |style="background:#ABCDEF;" align="center"| '''Product''' || DNA-Gyrase (subunit B) | |
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || DNA- | + | |- |
| − | |||
| − | |- | ||
| − | |||
|style="background:#ABCDEF;" align="center"|'''Function''' || DNA superrcoiling, initation of replication cycle and DNA elongation | |style="background:#ABCDEF;" align="center"|'''Function''' || DNA superrcoiling, initation of replication cycle and DNA elongation | ||
| − | |- | + | |- |
| − | |||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 71 kDa, 5.378 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 71 kDa, 5.378 | ||
| − | |- | + | |- |
| − | |||
|style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 1914 bp, 638 aa | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 1914 bp, 638 aa | ||
| − | |- | + | |- |
| − | |||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yaaB]]'', ''[[gyrA]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yaaB]]'', ''[[gyrA]]'' | ||
| − | |- | + | |- |
| − | + | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/gyrB_nucleotide.txt Gene sequence (+200bp) ]''' | |
| − | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni- | + | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/gyrB_protein.txt Protein sequence]''' |
| − | + | |- | |
| − | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni- | + | |colspan="2" | '''Genetic context''' <br/> [[Image:gyrB_context.gif]] |
| − | + | |- | |
| − | |- | ||
| − | |||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | ||
| − | |||
| − | |||
|} | |} | ||
| Line 58: | Line 43: | ||
=== Database entries === | === Database entries === | ||
| − | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yaaA- | + | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yaaA-recF-yaaB-gyrB.html] |
| − | |||
| − | |||
| − | |||
* '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10070] | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10070] | ||
| Line 90: | Line 72: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' [[GyrA]]- | + | * '''Interactions:''' [[GyrA]]-GyrB |
| − | |||
| − | * '''Localization:''' nucleoid- | + | * '''Localization:''' nucleoid-associated in actively growing cells [http://www.ncbi.nlm.nih.gov/sites/entrez/9539793 PubMed] |
| − | |||
=== Database entries === | === Database entries === | ||
| Line 102: | Line 82: | ||
* '''Swiss prot entry:''' | * '''Swiss prot entry:''' | ||
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget- | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU00060] |
| − | |||
* '''E.C. number:''' | * '''E.C. number:''' | ||
| Line 111: | Line 90: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''[[yaaA]]- | + | * '''Operon:''' ''[[yaaA]]-[[recF]]-[[yaaB]]-[[gyrB]]'' [http://www.ncbi.nlm.nih.gov/sites/entrez/2987848 PubMed] |
| − | |||
| − | |||
| − | |||
* '''Sigma factor:''' [[SigA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/2987848 PubMed] | * '''Sigma factor:''' [[SigA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/2987848 PubMed] | ||
| Line 122: | Line 98: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' GyrB is subject to Clp- | + | * '''Additional information:''' GyrB is subject to Clp-dependent proteolysis upon glucose starvation [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+17981983 PubMed] |
| − | |||
=Biological materials = | =Biological materials = | ||
| Line 135: | Line 110: | ||
* '''GFP fusion:''' | * '''GFP fusion:''' | ||
| − | * '''two- | + | * '''two-hybrid system:''' |
| − | |||
* '''Antibody:''' | * '''Antibody:''' | ||
| Line 146: | Line 120: | ||
=References= | =References= | ||
| − | # Huang et al. (1998) Bipolar localization of Bacillus subtilis topoisomerase IV, an enzyme required for chromosome segregation. ''Proc. Natl. Acad. Sci. USA'' '''95:''' 4652- | + | # Huang et al. (1998) Bipolar localization of Bacillus subtilis topoisomerase IV, an enzyme required for chromosome segregation. ''Proc. Natl. Acad. Sci. USA'' '''95:''' 4652-4657. [http://www.ncbi.nlm.nih.gov/sites/entrez/9539793 PubMed] |
| − | + | # Gerth et al. (2008) Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J Bacteriol 190:321-331 [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+17981983 PubMed] | |
| − | # Gerth et al. (2008) Clp- | + | # Ogasawara et al. (1985) Structure and function of the region of the replication origin of the ''Bacillus subtilis'' chromosome. IV. Transcription of the oriC region and expression of DNA gyrase genes and other open reading frames.''Nucl. Acids Res.'' '''11:''' 2267-2279. [http://www.ncbi.nlm.nih.gov/sites/entrez/2987848 PubMed] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | # Ogasawara et al. (1985) Structure and function of the region of the replication origin of the ''Bacillus subtilis'' chromosome. IV. Transcription of the oriC region and expression of DNA gyrase genes and other open reading frames.''Nucl. Acids Res.'' '''11:''' 2267- | ||
| − | |||
Revision as of 08:55, 23 March 2009
- Description: DNA-Gyrase (subunit B)
| Gene name | gyrB |
| Synonyms | novA |
| Essential | yes PubMed |
| Product | DNA-Gyrase (subunit B) |
| Function | DNA superrcoiling, initation of replication cycle and DNA elongation |
| MW, pI | 71 kDa, 5.378 |
| Gene length, protein length | 1914 bp, 638 aa |
| Immediate neighbours | yaaB, gyrA |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 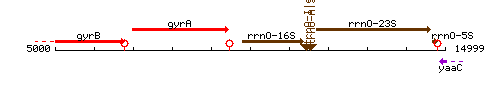
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): ParE
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions: GyrA-GyrB
- Localization: nucleoid-associated in actively growing cells PubMed
Database entries
- Structure:
- Swiss prot entry:
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information: GyrB is subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Huang et al. (1998) Bipolar localization of Bacillus subtilis topoisomerase IV, an enzyme required for chromosome segregation. Proc. Natl. Acad. Sci. USA 95: 4652-4657. PubMed
- Gerth et al. (2008) Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J Bacteriol 190:321-331 PubMed
- Ogasawara et al. (1985) Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. IV. Transcription of the oriC region and expression of DNA gyrase genes and other open reading frames.Nucl. Acids Res. 11: 2267-2279. PubMed