Difference between revisions of "YsxC"
(→Original publications) |
|||
| Line 156: | Line 156: | ||
==Original publications== | ==Original publications== | ||
| − | |||
<pubmed>17981968,11073929 ,12427945, 16997968 7961402 9852015 14684918 15136032 21636901 25771857</pubmed> | <pubmed>17981968,11073929 ,12427945, 16997968 7961402 9852015 14684918 15136032 21636901 25771857</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 07:27, 17 March 2015
- Description: assembly of the 50S subunit of the ribosome
| Gene name | ysxC |
| Synonyms | orfX |
| Essential | yes PubMed |
| Product | GTPase |
| Function | ribosome assembly |
| Gene expression levels in SubtiExpress: ysxC | |
| Interactions involving this protein in SubtInteract: YsxC | |
| Metabolic function and regulation of this protein in SubtiPathways: ysxC | |
| MW, pI | 21 kDa, 9.708 |
| Gene length, protein length | 585 bp, 195 aa |
| Immediate neighbours | ysxD, lonA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 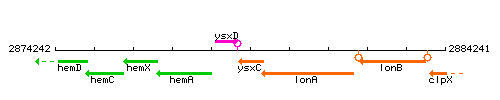
| |
Expression at a glance PubMed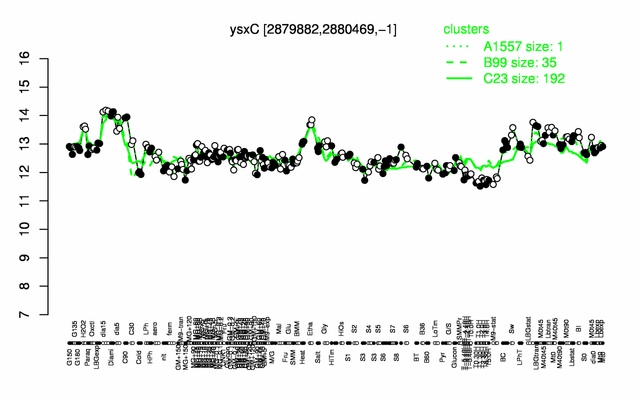
| |
Contents
Categories containing this gene/protein
translation, essential genes, membrane proteins, GTP-binding proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU28190
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU28190
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Binds and hydrolyzes GTP and readily exchanges GDP for GTP
- Protein family: engB family (according to Swiss-Prot) Era/Obg family
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- BsubCyc: BSU28190
- UniProt: P38424
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Naotake Ogasawara, Nara, Japan
Your additional remarks
References
Reviews
Natalie Verstraeten, Maarten Fauvart, Wim Versées, Jan Michiels
The universally conserved prokaryotic GTPases.
Microbiol Mol Biol Rev: 2011, 75(3);507-42, second and third pages of table of contents
[PubMed:21885683]
[WorldCat.org]
[DOI]
(I p)
Robert A Britton
Role of GTPases in bacterial ribosome assembly.
Annu Rev Microbiol: 2009, 63;155-76
[PubMed:19575570]
[WorldCat.org]
[DOI]
(I p)
Original publications
Catherine Wicker-Planquart, Jean-Michel Jault
Interaction between Bacillus subtilis YsxC and ribosomes (or rRNAs).
FEBS Lett: 2015, 589(9);1026-32
[PubMed:25771857]
[WorldCat.org]
[DOI]
(I p)
Kwok-Ho Chan, Kam-Bo Wong
Structure of an essential GTPase, YsxC, from Thermotoga maritima.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2011, 67(Pt 6);640-6
[PubMed:21636901]
[WorldCat.org]
[DOI]
(I p)
Catherine Wicker-Planquart, Anne-Emmanuelle Foucher, Mathilde Louwagie, Robert A Britton, Jean-Michel Jault
Interactions of an essential Bacillus subtilis GTPase, YsxC, with ribosomes.
J Bacteriol: 2008, 190(2);681-90
[PubMed:17981968]
[WorldCat.org]
[DOI]
(I p)
Laura Schaefer, William C Uicker, Catherine Wicker-Planquart, Anne-Emmanuelle Foucher, Jean-Michel Jault, Robert A Britton
Multiple GTPases participate in the assembly of the large ribosomal subunit in Bacillus subtilis.
J Bacteriol: 2006, 188(23);8252-8
[PubMed:16997968]
[WorldCat.org]
[DOI]
(P p)
Sergey N Ruzheinikov, Sanjan K Das, Svetlana E Sedelnikova, Patrick J Baker, Peter J Artymiuk, Jorge García-Lara, Simon J Foster, David W Rice
Analysis of the open and closed conformations of the GTP-binding protein YsxC from Bacillus subtilis.
J Mol Biol: 2004, 339(2);265-78
[PubMed:15136032]
[WorldCat.org]
[DOI]
(P p)
Sanjan K Das, Svetlana E Sedelnikova, Patrick J Baker, Sergey N Ruzheinikov, Simon J Foster, David W Rice
Expression, purification, crystallization and preliminary crystallographic analysis of a putative GTP-binding protein, YsxC, from Bacillus subtilis.
Acta Crystallogr D Biol Crystallogr: 2004, 60(Pt 1);166-8
[PubMed:14684918]
[WorldCat.org]
[DOI]
(P p)
Takuya Morimoto, Pek Chin Loh, Tomohiro Hirai, Kei Asai, Kazuo Kobayashi, Shigeki Moriya, Naotake Ogasawara
Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis.
Microbiology (Reading): 2002, 148(Pt 11);3539-3552
[PubMed:12427945]
[WorldCat.org]
[DOI]
(P p)
Z Prágai, C R Harwood
YsxC, a putative GTP-binding protein essential for growth of Bacillus subtilis 168.
J Bacteriol: 2000, 182(23);6819-23
[PubMed:11073929]
[WorldCat.org]
[DOI]
(P p)
E Krüger, M Hecker
The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes.
J Bacteriol: 1998, 180(24);6681-8
[PubMed:9852015]
[WorldCat.org]
[DOI]
(P p)
S Riethdorf, U Völker, U Gerth, A Winkler, S Engelmann, M Hecker
Cloning, nucleotide sequence, and expression of the Bacillus subtilis lon gene.
J Bacteriol: 1994, 176(21);6518-27
[PubMed:7961402]
[WorldCat.org]
[DOI]
(P p)