Difference between revisions of "GlnR"
(→Biological materials) |
|||
| Line 98: | Line 98: | ||
* '''Structure:''' | * '''Structure:''' | ||
| + | ** [[http://pdb.org/pdb/search/structidSearch.do?structureId=4r4e 4R4E]] (the GlnR-DNA complex) {{PubMed|25691471}} | ||
* '''UniProt:''' [http://www.uniprot.org/uniprot/P37582 P37582] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P37582 P37582] | ||
| Line 149: | Line 150: | ||
<pubmed>22625175 </pubmed> | <pubmed>22625175 </pubmed> | ||
==Original publications== | ==Original publications== | ||
| − | <pubmed>12374841,9287005,8799114, 18195355, 2573733, 8636055, 19233925 2906311 18331450 16547045 23535029</pubmed> | + | <pubmed>12374841,9287005,8799114, 18195355, 2573733, 8636055, 19233925 2906311 18331450 16547045 23535029 25691471</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:22, 19 February 2015
- Description: transcriptional repressor (MerR family) of the glnR-glnA operon
| Gene name | glnR |
| Synonyms | |
| Essential | no |
| Product | transcription repressor (MerR family) |
| Function | regulation of glutamine synthesis |
| Gene expression levels in SubtiExpress: glnR | |
| Interactions involving this protein in SubtInteract: GlnR | |
| Metabolic function and regulation of this protein in SubtiPathways: glnR | |
| MW, pI | 15 kDa, 9.731 |
| Gene length, protein length | 405 bp, 135 aa |
| Immediate neighbours | ynbB, glnA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 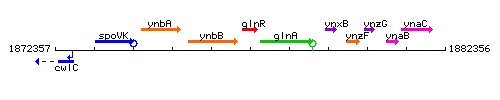
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids, glutamate metabolism, transcription factors and their control
This gene is a member of the following regulons
The GlnR regulon:
The gene
Basic information
- Locus tag: BSU17450
Phenotypes of a mutant
Database entries
- BsubCyc: BSU17450
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: MerR family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Effectors of protein activity: activity is enhanced upon interaction of the C-terminal domain with GlnA PubMed
Database entries
- BsubCyc: BSU17450
- UniProt: P37582
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: BP148 (del(glnR-glnA)::cat), available in Fabian Commichau's lab
- Expression vector:
- lacZ fusion: pGP189 (in pAC7), available in Jörg Stülke's lab
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Susan Fisher, Boston, USA homepage
Your additional remarks
References
Reviews
Original publications
Maria A Schumacher, Naga Babu Chinnam, Bonnie Cuthbert, Nam K Tonthat, Travis Whitfill
Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev: 2015, 29(4);451-64
[PubMed:25691471]
[WorldCat.org]
[DOI]
(I p)
Ksenia Fedorova, Airat Kayumov, Kathrin Woyda, Olga Ilinskaja, Karl Forchhammer
Transcription factor TnrA inhibits the biosynthetic activity of glutamine synthetase in Bacillus subtilis.
FEBS Lett: 2013, 587(9);1293-8
[PubMed:23535029]
[WorldCat.org]
[DOI]
(I p)
Susan H Fisher, Lewis V Wray
Novel trans-Acting Bacillus subtilis glnA mutations that derepress glnRA expression.
J Bacteriol: 2009, 191(8);2485-92
[PubMed:19233925]
[WorldCat.org]
[DOI]
(I p)
Lewis V Wray, Susan H Fisher
Bacillus subtilis GlnR contains an autoinhibitory C-terminal domain required for the interaction with glutamine synthetase.
Mol Microbiol: 2008, 68(2);277-85
[PubMed:18331450]
[WorldCat.org]
[DOI]
(I p)
Susan H Fisher, Lewis V Wray
Bacillus subtilis glutamine synthetase regulates its own synthesis by acting as a chaperone to stabilize GlnR-DNA complexes.
Proc Natl Acad Sci U S A: 2008, 105(3);1014-9
[PubMed:18195355]
[WorldCat.org]
[DOI]
(I p)
Jill M Zalieckas, Lewis V Wray, Susan H Fisher
Cross-regulation of the Bacillus subtilis glnRA and tnrA genes provides evidence for DNA binding site discrimination by GlnR and TnrA.
J Bacteriol: 2006, 188(7);2578-85
[PubMed:16547045]
[WorldCat.org]
[DOI]
(P p)
Jaclyn L Brandenburg, Lewis V Wray, Lars Beier, Hanne Jarmer, Hans H Saxild, Susan H Fisher
Roles of PucR, GlnR, and TnrA in regulating expression of the Bacillus subtilis ure P3 promoter.
J Bacteriol: 2002, 184(21);6060-4
[PubMed:12374841]
[WorldCat.org]
[DOI]
(P p)
L V Wray, A E Ferson, S H Fisher
Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H.
J Bacteriol: 1997, 179(17);5494-501
[PubMed:9287005]
[WorldCat.org]
[DOI]
(P p)
L V Wray, A E Ferson, K Rohrer, S H Fisher
TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis.
Proc Natl Acad Sci U S A: 1996, 93(17);8841-5
[PubMed:8799114]
[WorldCat.org]
[DOI]
(P p)
S W Brown, A L Sonenshein
Autogenous regulation of the Bacillus subtilis glnRA operon.
J Bacteriol: 1996, 178(8);2450-4
[PubMed:8636055]
[WorldCat.org]
[DOI]
(P p)
H J Schreier, S W Brown, K D Hirschi, J F Nomellini, A L Sonenshein
Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene.
J Mol Biol: 1989, 210(1);51-63
[PubMed:2573733]
[WorldCat.org]
[DOI]
(P p)
M A Strauch, A I Aronson, S W Brown, H J Schreier, A L Sonenhein
Sequence of the Bacillus subtilis glutamine synthetase gene region.
Gene: 1988, 71(2);257-65
[PubMed:2906311]
[WorldCat.org]
[DOI]
(P p)