Difference between revisions of "YjbH"
(→Original Publications) |
|||
| Line 85: | Line 85: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| + | ** [[YjbH]] aggregates under stress conditions, resulting in the inability to bind to [[Spx]], and therefore in stabilization of [[Spx]] {{PubMed|25353645}} | ||
** Zn atom is released upon treatment with strong oxidants [http://www.ncbi.nlm.nih.gov/sites/entrez/19074380 PubMed] | ** Zn atom is released upon treatment with strong oxidants [http://www.ncbi.nlm.nih.gov/sites/entrez/19074380 PubMed] | ||
** interaction with [[YirB]] inhibits the formation of a complex with [[Spx]] {{PubMed|21378193}} | ** interaction with [[YirB]] inhibits the formation of a complex with [[Spx]] {{PubMed|21378193}} | ||
| Line 149: | Line 150: | ||
<pubmed> 23375660 23479438,19609260</pubmed> | <pubmed> 23375660 23479438,19609260</pubmed> | ||
==Original Publications== | ==Original Publications== | ||
| − | <pubmed>17293416, 19074380 17908206, 20525796 21378193,21947404 24417481 24942655 </pubmed> | + | <pubmed>17293416, 19074380 17908206, 20525796 21378193,21947404 24417481 24942655 25353645 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:28, 31 October 2014
- Description: adaptor protein for ClpX-ClpP-catalyzed Spx degradation, confers resistance against nitrosating agents
| Gene name | yjbH |
| Synonyms | |
| Essential | no |
| Product | adaptor protein |
| Function | stimulation of Spx degradation |
| Gene expression levels in SubtiExpress: yjbH | |
| Interactions involving this protein in SubtInteract: YjbH | |
| MW, pI | 31 kDa, 5.206 |
| Gene length, protein length | 825 bp, 275 aa |
| Immediate neighbours | yizD, yjbI |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed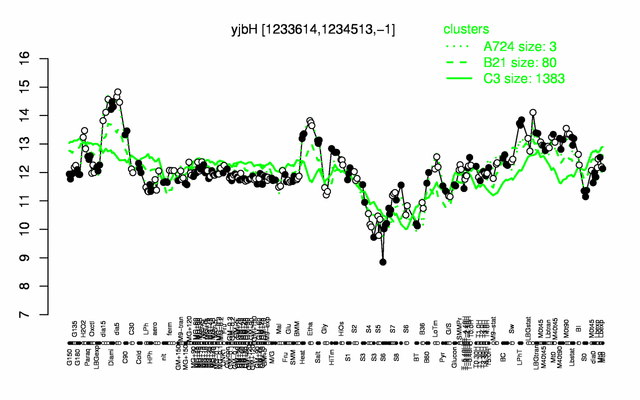
| |
Contents
Categories containing this gene/protein
proteolysis, resistance against other toxic compounds (nitric oxide, phenolic acids, flavonoids, oxalate)
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU11550
Phenotypes of a mutant
- increased thermotolerance due to increased stabiliy of Spx and thus increased expression of trxA PubMed
Database entries
- BsubCyc: BSU11550
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: adaptor protein for ClpX-ClpP-catalyzed Spx degradation PubMed
- Protein family: UPF0413 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization: cytosolic protein
Database entries
- BsubCyc: BSU11550
- Structure:
- UniProt: O31606
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Peter Zuber, Oregon Health and Science University, USA Homepage
Claes von Wachenfeldt, Lund University, Sweden Homepage
Your additional remarks
References
Reviews
Original Publications
Jakob Engman, Claes von Wachenfeldt
Regulated protein aggregation: a mechanism to control the activity of the ClpXP adaptor protein YjbH.
Mol Microbiol: 2015, 95(1);51-63
[PubMed:25353645]
[WorldCat.org]
[DOI]
(I p)
Chio Mui Chan, Erik Hahn, Peter Zuber
Adaptor bypass mutations of Bacillus subtilis spx suggest a mechanism for YjbH-enhanced proteolysis of the regulator Spx by ClpXP.
Mol Microbiol: 2014, 93(3);426-38
[PubMed:24942655]
[WorldCat.org]
[DOI]
(I p)
Stephanie Runde, Noël Molière, Anja Heinz, Etienne Maisonneuve, Armgard Janczikowski, Alexander K W Elsholz, Ulf Gerth, Michael Hecker, Kürşad Turgay
The role of thiol oxidative stress response in heat-induced protein aggregate formation during thermotolerance in Bacillus subtilis.
Mol Microbiol: 2014, 91(5);1036-52
[PubMed:24417481]
[WorldCat.org]
[DOI]
(I p)
Nadine Göhring, Iris Fedtke, Guoqing Xia, Ana M Jorge, Mariana G Pinho, Ute Bertsche, Andreas Peschel
New role of the disulfide stress effector YjbH in β-lactam susceptibility of Staphylococcus aureus.
Antimicrob Agents Chemother: 2011, 55(12);5452-8
[PubMed:21947404]
[WorldCat.org]
[DOI]
(I p)
Sushma Kommineni, Saurabh K Garg, Chio Mui Chan, Peter Zuber
YjbH-enhanced proteolysis of Spx by ClpXP in Bacillus subtilis is inhibited by the small protein YirB (YuzO).
J Bacteriol: 2011, 193(9);2133-40
[PubMed:21378193]
[WorldCat.org]
[DOI]
(I p)
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Saurabh K Garg, Sushma Kommineni, Luke Henslee, Ying Zhang, Peter Zuber
The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx.
J Bacteriol: 2009, 191(4);1268-77
[PubMed:19074380]
[WorldCat.org]
[DOI]
(I p)
Jonas T Larsson, Annika Rogstam, Claes von Wachenfeldt
YjbH is a novel negative effector of the disulphide stress regulator, Spx, in Bacillus subtilis.
Mol Microbiol: 2007, 66(3);669-84
[PubMed:17908206]
[WorldCat.org]
[DOI]
(P p)
Annika Rogstam, Jonas T Larsson, Peter Kjelgaard, Claes von Wachenfeldt
Mechanisms of adaptation to nitrosative stress in Bacillus subtilis.
J Bacteriol: 2007, 189(8);3063-71
[PubMed:17293416]
[WorldCat.org]
[DOI]
(P p)