Difference between revisions of "AbrB"
(→Other original publications) |
(→Other original publications) |
||
| Line 160: | Line 160: | ||
<pubmed>20817675,18840696 23580539</pubmed> | <pubmed>20817675,18840696 23580539</pubmed> | ||
==Other original publications== | ==Other original publications== | ||
| − | <pubmed> 19581368 ,3145384,2437099,8878039,2504584, 11377867,1850083,2106683,12586407, 12076816,17660417, 2554317,18430133,7768874, 2507867, 1766371, 15687200,1908787,2106683,15687200, 2504584,3145384 8821944 8576231 11101897 11395475 11583849 11751836 12123659 12076816 12591885 15610005 16223496 16159768 16702211 17660417 17720793 7592460, 19465659 20509597 15101989 19202088,18326573 24534728 24731262 24832089 </pubmed> | + | <pubmed> 19581368 ,3145384,2437099,8878039,2504584, 11377867,1850083,2106683,12586407, 12076816,17660417, 2554317,18430133,7768874, 2507867, 1766371, 15687200,1908787,2106683,15687200, 2504584,3145384 8821944 8576231 11101897 11395475 11583849 11751836 12123659 12076816 12591885 15610005 16223496 16159768 16702211 17660417 17720793 7592460, 19465659 20509597 15101989 19202088,18326573 24534728 24731262 24832089 25002359 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:27, 10 July 2014
- Description: transcriptional regulator of transition state genes
| Gene name | abrB |
| Synonyms | cpsX, tolB |
| Essential | no |
| Product | transcriptional regulator |
| Function | regulation of gene expression during the transition from growth to stationary phase |
| Gene expression levels in SubtiExpress: abrB | |
| Interactions involving this protein in SubtInteract: AbrB | |
| Metabolic function and regulation of this protein in SubtiPathways: abrB | |
| MW, pI | 10 kDa, 6.57 |
| Gene length, protein length | 282 bp, 94 aa |
| Immediate neighbours | yabC, metS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 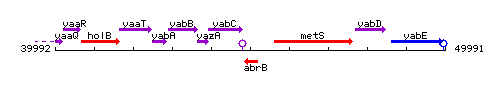
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The AbrB regulon
The gene
Basic information
- Locus tag: BSU00370
Phenotypes of a mutant
- No swarming motility on B medium PubMed
Database entries
- BsubCyc: BSU00370
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
Extended information on the protein
- Kinetic information:
- Modification: phosphorylated on Ser-86 PubMed by PrkC, PrkD, and YabT results in loss of DNA-binding activity PubMed
Database entries
- BsubCyc: BSU00370
- UniProt: P08874
- KEGG entry: [3]
Additional information
Expression and regulation
- Operon: abrB PubMed
- Regulation: expressed at the onset of stationary phase PubMed
- Additional information:
Biological materials
- Mutant: TT731 (aphA3)
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Richard Losick, Harvard Univ., Cambridge, USA homepage
Mark Strauch, Baltimore, USA homepage
Your additional remarks
References
Reviews
The AbrB regulon:
Other original publications