Difference between revisions of "AdhR"
| Line 1: | Line 1: | ||
| − | * '''Description:''' MerR | + | * '''Description:''' transcriptional activator ([[transcription factors of the MerR family|MerR family]]) of ''[[adhA]]-[[yraA]]'', responsive to formaldehyde and methylglyoxal <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || MerR | + | |style="background:#ABCDEF;" align="center"| '''Product''' || transcriptional activator ([[transcription factors of the MerR family|MerR family]]) |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || regulation of the protective response to formaldehyde and methylglyoxal | |style="background:#ABCDEF;" align="center"|'''Function''' || regulation of the protective response to formaldehyde and methylglyoxal | ||
| Line 37: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 66: | Line 62: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 76: | Line 69: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| − | * '''Protein family:'''MerR | + | * '''Protein family:''' [[transcription factors of the MerR family|MerR family]] |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
Latest revision as of 13:14, 22 June 2014
- Description: transcriptional activator (MerR family) of adhA-yraA, responsive to formaldehyde and methylglyoxal
| Gene name | adhR |
| Synonyms | yraB |
| Essential | no |
| Product | transcriptional activator (MerR family) |
| Function | regulation of the protective response to formaldehyde and methylglyoxal |
| Gene expression levels in SubtiExpress: adhR | |
| MW, pI | 16 kDa, 9.637 |
| Gene length, protein length | 420 bp, 140 aa |
| Immediate neighbours | yraD, yrzP |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 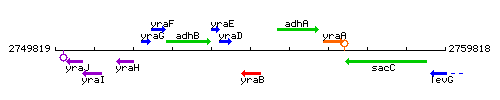
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed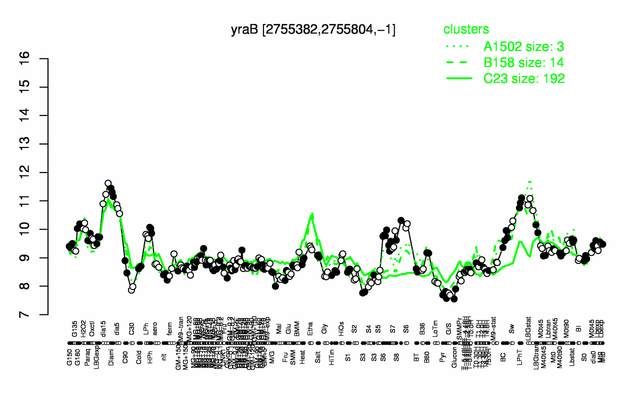
| |
Contents
Categories containing this gene/protein
transcription factors and their control, resistance against oxidative and electrophile stress
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU27000
Phenotypes of a mutant
Database entries
- BsubCyc: BSU27000
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: MerR family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: activity probably redox-controlled via thiol-(S)-alkylation at Cys-52 by aldehydes PubMed
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasmic
Database entries
- BsubCyc: BSU27000
- Structure:
- UniProt: O06008
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Haike Antelmann,University of Greifswald, Germany
Your additional remarks
References
Reviews
Antelmann H, Helmann JD. Thiol-based redox switches and gene regulation. Antioxid Redox Signal. 2011,14:1049-63. PubMed
Original articles
Thi Thu Huyen Nguyen, Warawan Eiamphungporn, Ulrike Mäder, Manuel Liebeke, Michael Lalk, Michael Hecker, John D Helmann, Haike Antelmann
Genome-wide responses to carbonyl electrophiles in Bacillus subtilis: control of the thiol-dependent formaldehyde dehydrogenase AdhA and cysteine proteinase YraA by the MerR-family regulator YraB (AdhR).
Mol Microbiol: 2009, 71(4);876-94
[PubMed:19170879]
[WorldCat.org]
[DOI]
(I p)