Difference between revisions of "NsrR"
(→Reviews) |
(→Other original publications) |
||
| Line 143: | Line 143: | ||
<pubmed> 22287527 </pubmed> | <pubmed> 22287527 </pubmed> | ||
==Other original publications== | ==Other original publications== | ||
| − | <pubmed>16885456, 24214949,17293416,21091510 , 19006327</pubmed> | + | <pubmed>16885456, 24214949,17293416,21091510 , 19006327 18989365 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 15:40, 26 May 2014
- Description: nitric oxide-responsive regulator
| Gene name | nsrR |
| Synonyms | yhdE |
| Essential | no |
| Product | transcriptional repressor |
| Function | repression of ResD-ResE-dependent genes in the absence of nitric oxide (NO) |
| Gene expression levels in SubtiExpress: nsrR | |
| Metabolic function and regulation of this protein in SubtiPathways: nsrR | |
| MW, pI | 16 kDa, 7.75 |
| Gene length, protein length | 438 bp, 146 aa |
| Immediate neighbours | lytF, ygxB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 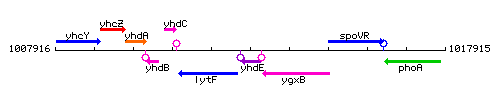
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed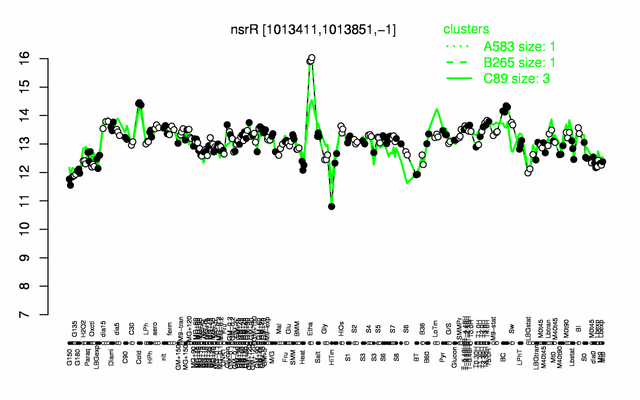
| |
Contents
Categories containing this gene/protein
transcription factors and their control, resistance against other toxic compounds (nitric oxide, phenolic acids, flavonoids, oxalate)
This gene is a member of the following regulons
The NsrR regulon
The gene
Basic information
- Locus tag: BSU09380
Phenotypes of a mutant
Database entries
- BsubCyc: BSU09380
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): [4Fe-4S] cluster, required for DNA-binding PubMed
- Effectors of protein activity:
Database entries
- BsubCyc: BSU09380
- Structure:
- UniProt: O07573
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: nsrR PubMed
- Regulation:
- Regulatory mechanism:
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium): 122 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
The NsrR regulon
Other original publications