Difference between revisions of "PdhA"
| Line 1: | Line 1: | ||
| − | * '''Description:''' pyruvate dehydrogenase (E1 alpha subunit)<br/><br/> | + | * '''Description:''' pyruvate dehydrogenase (E1 alpha subunit), required for Z-ring assembly in a pyruvate-dependent manner<br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 8: | Line 8: | ||
|style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''aceA '' | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''aceA '' | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Essential''' || yes | + | |style="background:#ABCDEF;" align="center"| '''Essential''' || yes {{PubMed|22383849}}, no {{PubMed|24825009}} |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Product''' || pyruvate dehydrogenase (E1 alpha subunit) | |style="background:#ABCDEF;" align="center"| '''Product''' || pyruvate dehydrogenase (E1 alpha subunit) | ||
| Line 57: | Line 57: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | * ''pdhA'' is essential [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed] | + | * ''pdhA'' is essential according to Kobayashi ''et al''. [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed] |
| + | * the mutant grows slowly but is viable {{PubMed|24825009}} | ||
| + | * depletion of ''[[pdhA]]'' and deletion of ''[[ezrA]]'' have a strong synthetic defect in [[cell division]] {{PubMed|24825009}} | ||
=== Database entries === | === Database entries === | ||
| Line 97: | Line 99: | ||
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
| + | ** colocalizes with the nucleoid (depending on the availability of pyruvate) {{PubMed|24825009}} | ||
=== Database entries === | === Database entries === | ||
| Line 158: | Line 161: | ||
<pubmed> 19476487 9655937 2227213 6805383 </pubmed> | <pubmed> 19476487 9655937 2227213 6805383 </pubmed> | ||
==Original publications== | ==Original publications== | ||
| − | <pubmed>9352926, 20525796, 12850135 6414463 11976308 20081037 15378759</pubmed> | + | <pubmed>9352926, 20525796, 12850135 6414463 11976308 20081037 15378759 24825009</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 17:03, 16 May 2014
- Description: pyruvate dehydrogenase (E1 alpha subunit), required for Z-ring assembly in a pyruvate-dependent manner
| Gene name | pdhA |
| Synonyms | aceA |
| Essential | yes PubMed, no PubMed |
| Product | pyruvate dehydrogenase (E1 alpha subunit) |
| Function | links glycolysis and TCA cycle |
| Gene expression levels in SubtiExpress: pdhA | |
| Interactions involving this protein in SubtInteract: PdhA | |
| Metabolic function and regulation of this protein in SubtiPathways: pdhA | |
| MW, pI | 41 kDa, 5.837 |
| Gene length, protein length | 1113 bp, 371 aa |
| Immediate neighbours | ykyA, pdhB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 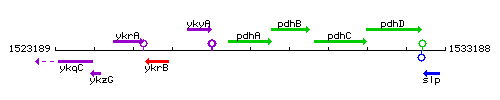
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
carbon core metabolism, essential genes, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU14580
Phenotypes of a mutant
- pdhA is essential according to Kobayashi et al. PubMed
- the mutant grows slowly but is viable PubMed
- depletion of pdhA and deletion of ezrA have a strong synthetic defect in cell division PubMed
Database entries
- BsubCyc: BSU14580
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Pyruvate + [dihydrolipoyllysine-residue acetyltransferase] lipoyllysine = [dihydrolipoyllysine-residue acetyltransferase] S-acetyldihydrolipoyllysine + CO2 (according to Swiss-Prot)
- Protein family:
Extended information on the protein
- Kinetic information: Michaelis-Menten PubMed
- Modification:
- Cofactors:
- thiamine pyrophosphate
- Effectors of protein activity:
- Inhibited thiamine 2-thiothiazolone diphosphate and NADH PubMed
- Low sensibility to NADPH
- Localization:
- colocalizes with the nucleoid (depending on the availability of pyruvate) PubMed
Database entries
- BsubCyc: BSU14580
- Structure: 1W88 (E1 in complex with subunit binding domain of E2, Geobacillus stearothermophilus)
- UniProt: P21881
- KEGG entry: [3]
- E.C. number: 1.2.4.1
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- stringent response: due to presence of guanine at +1 position of the transcript PubMed
- Additional information:
- The mRNA has a long 5' leader region. This may indicate RNA-based regulation PubMed
- belongs to the 100 most abundant proteins PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 5117 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 18311 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 4425 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 5452 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 7055 PubMed
Biological materials
- Mutant:
- Expression vector:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Arthur Aronson, Purdue University, West Lafayette, USA homepage
Your additional remarks
References
Reviews
Original publications