Difference between revisions of "AcsA"
m (Reverted edits by 134.76.70.252 (talk) to last revision by 134.76.38.147) |
|||
| Line 126: | Line 126: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| − | |||
| − | |||
| − | |||
| − | + | =Biological materials = | |
| − | |||
* '''Mutant:''' | * '''Mutant:''' | ||
** GP1212 (''acsA''::''kan''), available in [[Stülke]] lab | ** GP1212 (''acsA''::''kan''), available in [[Stülke]] lab | ||
Revision as of 13:42, 17 April 2014
- Description: acetyl-CoA synthetase
| Gene name | acsA |
| Synonyms | |
| Essential | no |
| Product | acetyl-CoA synthetase) |
| Function | utilization of acetate, fatty acids |
| Gene expression levels in SubtiExpress: acsA | |
| Metabolic function and regulation of this protein in SubtiPathways: acsA | |
| MW, pI | 64 kDa, 5.547 |
| Gene length, protein length | 1716 bp, 572 aa |
| Immediate neighbours | ytzK, acuA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 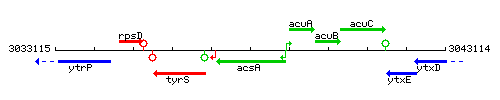
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed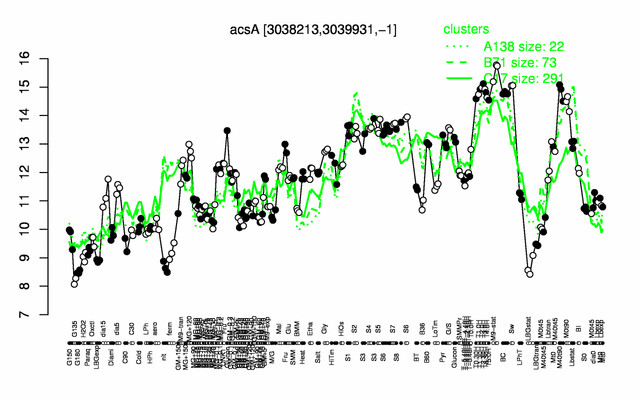
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources, utilization of lipids
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29680
Phenotypes of a mutant
Database entries
- BsubCyc: BSU29680
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + acetate + CoA = AMP + diphosphate + acetyl-CoA (according to Swiss-Prot)
- Protein family: ATP-dependent AMP-binding enzyme family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: acetylated on Lys-549 by AcuA, this results in inactivation PubMed, deacetylated by SrtN and AcuC deacetylates (and thereby activates) AcsA PubMed
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU29680
- Structure:
- UniProt: P39062
- KEGG entry: [3]
- E.C. number: 6.2.1.1
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- GP1212 (acsA::kan), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Eric L Hegg
Unraveling the structure and mechanism of acetyl-coenzyme A synthase.
Acc Chem Res: 2004, 37(10);775-83
[PubMed:15491124]
[WorldCat.org]
[DOI]
(P p)
V J Starai, J C Escalante-Semerena
Acetyl-coenzyme A synthetase (AMP forming).
Cell Mol Life Sci: 2004, 61(16);2020-30
[PubMed:15316652]
[WorldCat.org]
[DOI]
(P p)
Paul A Lindahl
Acetyl-coenzyme A synthase: the case for a Ni(p)(0)-based mechanism of catalysis.
J Biol Inorg Chem: 2004, 9(5);516-24
[PubMed:15221478]
[WorldCat.org]
[DOI]
(P p)
Original publications