Difference between revisions of "InfB"
| Line 59: | Line 59: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU16630&redirect=T BSU16630] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ylxS-nusA-ylxRQ-infB-ylxP-rbfA.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ylxS-nusA-ylxRQ-infB-ylxP-rbfA.html] | ||
| Line 96: | Line 97: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU16630&redirect=T BSU16630] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1D1N 1D1N] (fMet-tRNA-fMet binding domain, Geobacillus stearothermophilus), [http://www.rcsb.org/pdb/explore.do?structureId=1Z9B 1Z9B] (C1-subdomain, Geobacillus stearothermophilus) | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1D1N 1D1N] (fMet-tRNA-fMet binding domain, Geobacillus stearothermophilus), [http://www.rcsb.org/pdb/explore.do?structureId=1Z9B 1Z9B] (C1-subdomain, Geobacillus stearothermophilus) | ||
Revision as of 13:45, 2 April 2014
- Description: translation initiation factor IF-2
| Gene name | infB |
| Synonyms | |
| Essential | yes PubMed |
| Product | initiation factor IF-2 |
| Function | translation |
| Gene expression levels in SubtiExpress: infB | |
| MW, pI | 78 kDa, 5.191 |
| Gene length, protein length | 2148 bp, 716 aa |
| Immediate neighbours | ylxQ, ylxP |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 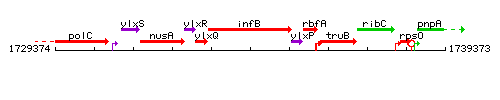
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed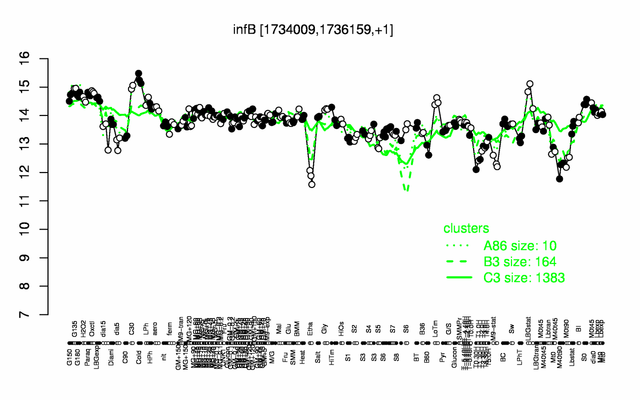
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU16630
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU16630
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Initiation of translation
- Protein family: IF-2 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains: Nucleotide-binding domain that competitively binds GTP or ppGpp PubMed
- Modification:
- Cofactor(s):
- Effectors of protein activity: GTP and ppGpp PubMed
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU16630
- Structure: 1D1N (fMet-tRNA-fMet binding domain, Geobacillus stearothermophilus), 1Z9B (C1-subdomain, Geobacillus stearothermophilus)
- UniProt: P17889
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information: Putative cellular metabolic sensor and regulator that oscillates between an active GTP-bound form under conditions allowing active protein syntheses and an inactive ppGpp-bound form when depleting of nutrients PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Pohl Milon, Eugene Tischenko, Jerneja Tomsic, Enrico Caserta, Gert Folkers, Anna La Teana, Marina V Rodnina, Cynthia L Pon, Rolf Boelens, Claudio O Gualerzi
The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor.
Proc Natl Acad Sci U S A: 2006, 103(38);13962-7
[PubMed:16968770]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Georg Homuth, Christian Scharf, Michael Hecker
Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis.
J Bacteriol: 2002, 184(9);2500-20
[PubMed:11948165]
[WorldCat.org]
[DOI]
(P p)
K Shazand, J Tucker, M Grunberg-Manago, J C Rabinowitz, T Leighton
Similar organization of the nusA-infB operon in Bacillus subtilis and Escherichia coli.
J Bacteriol: 1993, 175(10);2880-7
[PubMed:8491709]
[WorldCat.org]
[DOI]
(P p)