Difference between revisions of "RsbS"
| Line 80: | Line 80: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' phosphorylation on Ser-59 by [[RsbT]] [http://www.ncbi.nlm.nih.gov/sites/entrez/17218307 PubMed], dephosphorylation by [[RsbX]] {{PubMed|21362065}} | * '''Modification:''' phosphorylation on Ser-59 by [[RsbT]] [http://www.ncbi.nlm.nih.gov/sites/entrez/17218307 PubMed], dephosphorylation by [[RsbX]] {{PubMed|21362065}} | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 148: | Line 148: | ||
==Reviews== | ==Reviews== | ||
<pubmed>19704888 16319496 20658979 </pubmed> | <pubmed>19704888 16319496 20658979 </pubmed> | ||
| − | |||
==Original Articles== | ==Original Articles== | ||
| − | + | <pubmed>8002610,8682769,9786195,8682789,10781545, 15583165,8824586,10329124,17158665, 21821766 16321960,10671474,8808936,15312768,11244072,15342582,,16321960, 8955331, 12950928, 15466036, 9179850, 18832644 ,17218307 20019076 24599254 23320651,21362065</pubmed> | |
| − | <pubmed>8002610,8682769,9786195,8682789,10781545, 15583165,8824586,10329124,17158665, 21821766 16321960,10671474,8808936,15312768,11244072,15342582,,16321960, 8955331, 12950928, 15466036, 9179850, 18832644 ,17218307 20019076</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 17:27, 8 March 2014
- Description: scaffold protein of the stressosome, anti-RsbT
| Gene name | rsbS |
| Synonyms | ycxS |
| Essential | no |
| Product | scaffold protein of the stressosome, anti-RsbT |
| Function | control of SigB activity |
| Gene expression levels in SubtiExpress: rsbS | |
| Interactions involving this protein in SubtInteract: RsbS | |
| Metabolic function and regulation of this protein in SubtiPathways: rsbS | |
| MW, pI | 13 kDa, 4.14 |
| Gene length, protein length | 363 bp, 121 aa |
| Immediate neighbours | rsbR, rsbT |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 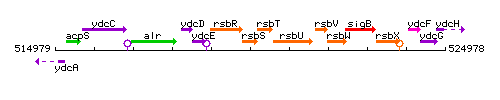
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed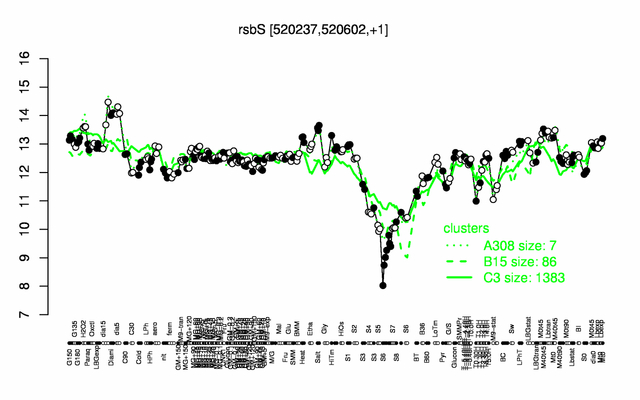
| |
Contents
Categories containing this gene/protein
sigma factors and their control, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU04680
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Effectors of protein activity:
Database entries
- Structure: 3VY9 (complete stressosome)
- UniProt: P42410
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation: constitutively expressed PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Bill Haldenwang, San Antonio, USA
- Chet Price, Davis, USA homepage
- Rick Lewis, Newcastle, UK homepage
Your additional remarks
References
Reviews
Jon Marles-Wright, Richard J Lewis
The stressosome: molecular architecture of a signalling hub.
Biochem Soc Trans: 2010, 38(4);928-33
[PubMed:20658979]
[WorldCat.org]
[DOI]
(I p)
Jon Marles-Wright, Richard J Lewis
The Bacillus subtilis stressosome: A signal integration and transduction hub.
Commun Integr Biol: 2008, 1(2);182-4
[PubMed:19704888]
[WorldCat.org]
[DOI]
(I p)
Jan Pané-Farré, Richard J Lewis, Jörg Stülke
The RsbRST stress module in bacteria: a signalling system that may interact with different output modules.
J Mol Microbiol Biotechnol: 2005, 9(2);65-76
[PubMed:16319496]
[WorldCat.org]
[DOI]
(P p)
Original Articles
Tatiana A Gaidenko, Chester W Price
Genetic evidence for a phosphorylation-independent signal transduction mechanism within the Bacillus subtilis stressosome.
PLoS One: 2014, 9(3);e90741
[PubMed:24599254]
[WorldCat.org]
[DOI]
(I e)
Ulf W Liebal, Thomas Millat, Jon Marles-Wright, Richard J Lewis, Olaf Wolkenhauer
Simulations of stressosome activation emphasize allosteric interactions between RsbR and RsbT.
BMC Syst Biol: 2013, 7;3
[PubMed:23320651]
[WorldCat.org]
[DOI]
(I e)
Christopher T Brown, Laura K Fishwick, Binna M Chokshi, Marissa A Cuff, Jay M Jackson, Travis Oglesby, Alison T Rioux, Enrique Rodriguez, Gregory S Stupp, Austin H Trupp, James S Woollcombe-Clarke, Tracy N Wright, William J Zaragoza, Jennifer C Drew, Eric W Triplett, Wayne L Nicholson
Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation.
Appl Environ Microbiol: 2011, 77(19);6867-77
[PubMed:21821766]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Stephan Schulz, Katrin Gronau, Dörte Becher, Michael Hecker, Chester W Price
In vivo phosphorylation patterns of key stressosome proteins define a second feedback loop that limits activation of Bacillus subtilis σB.
Mol Microbiol: 2011, 80(3);798-810
[PubMed:21362065]
[WorldCat.org]
[DOI]
(I p)
Adam Reeves, Luis Martinez, William Haldenwang
Expression of, and in vivo stressosome formation by, single members of the RsbR protein family in Bacillus subtilis.
Microbiology (Reading): 2010, 156(Pt 4);990-998
[PubMed:20019076]
[WorldCat.org]
[DOI]
(I p)
Jon Marles-Wright, Tim Grant, Olivier Delumeau, Gijs van Duinen, Susan J Firbank, Peter J Lewis, James W Murray, Joseph A Newman, Maureen B Quin, Paul R Race, Alexis Rohou, Willem Tichelaar, Marin van Heel, Richard J Lewis
Molecular architecture of the "stressosome," a signal integration and transduction hub.
Science: 2008, 322(5898);92-6
[PubMed:18832644]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Adam Reeves, W G Haldenwang
Isolation and characterization of dominant mutations in the Bacillus subtilis stressosome components RsbR and RsbS.
J Bacteriol: 2007, 189(5);1531-41
[PubMed:17158665]
[WorldCat.org]
[DOI]
(P p)
Shuyu Zhang, Adam Reeves, Robyn L Woodbury, W G Haldenwang
Coexpression patterns of sigma(B) regulators in Bacillus subtilis affect sigma(B) inducibility.
J Bacteriol: 2005, 187(24);8520-5
[PubMed:16321960]
[WorldCat.org]
[DOI]
(P p)
Shrin Kuo, Shuyu Zhang, Robyn L Woodbury, W G Haldenwang
Associations between Bacillus subtilis sigmaB regulators in cell extracts.
Microbiology (Reading): 2004, 150(Pt 12);4125-36
[PubMed:15583165]
[WorldCat.org]
[DOI]
(P p)
Chien-Cheng Chen, Michael D Yudkin, Olivier Delumeau
Phosphorylation and RsbX-dependent dephosphorylation of RsbR in the RsbR-RsbS complex of Bacillus subtilis.
J Bacteriol: 2004, 186(20);6830-6
[PubMed:15466036]
[WorldCat.org]
[DOI]
(P p)
Tae-Jong Kim, Tatiana A Gaidenko, Chester W Price
In vivo phosphorylation of partner switching regulators correlates with stress transmission in the environmental signaling pathway of Bacillus subtilis.
J Bacteriol: 2004, 186(18);6124-32
[PubMed:15342582]
[WorldCat.org]
[DOI]
(P p)
Tae-Jong Kim, Tatiana A Gaidenko, Chester W Price
A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis.
J Mol Biol: 2004, 341(1);135-50
[PubMed:15312768]
[WorldCat.org]
[DOI]
(P p)
Chien-Cheng Chen, Richard J Lewis, Robin Harris, Michael D Yudkin, Olivier Delumeau
A supramolecular complex in the environmental stress signalling pathway of Bacillus subtilis.
Mol Microbiol: 2003, 49(6);1657-69
[PubMed:12950928]
[WorldCat.org]
[DOI]
(P p)
S Zhang, J M Scott, W G Haldenwang
Loss of ribosomal protein L11 blocks stress activation of the Bacillus subtilis transcription factor sigma(B).
J Bacteriol: 2001, 183(7);2316-21
[PubMed:11244072]
[WorldCat.org]
[DOI]
(P p)
J M Scott, J Ju, T Mitchell, W G Haldenwang
The Bacillus subtilis GTP binding protein obg and regulators of the sigma(B) stress response transcription factor cofractionate with ribosomes.
J Bacteriol: 2000, 182(10);2771-7
[PubMed:10781545]
[WorldCat.org]
[DOI]
(P p)
J M Scott, T Mitchell, W G Haldenwang
Stress triggers a process that limits activation of the Bacillus subtilis stress transcription factor sigma(B).
J Bacteriol: 2000, 182(5);1452-6
[PubMed:10671474]
[WorldCat.org]
[DOI]
(P p)
T A Gaidenko, X Yang, Y M Lee, C W Price
Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the environmental stress signaling pathway of Bacillus subtilis.
J Mol Biol: 1999, 288(1);29-39
[PubMed:10329124]
[WorldCat.org]
[DOI]
(P p)
C M Kang, K Vijay, C W Price
Serine kinase activity of a Bacillus subtilis switch protein is required to transduce environmental stress signals but not to activate its target PP2C phosphatase.
Mol Microbiol: 1998, 30(1);189-96
[PubMed:9786195]
[WorldCat.org]
[DOI]
(P p)
S Akbar, C M Kang, T A Gaidenko, C W Price
Modulator protein RsbR regulates environmental signalling in the general stress pathway of Bacillus subtilis.
Mol Microbiol: 1997, 24(3);567-78
[PubMed:9179850]
[WorldCat.org]
[DOI]
(P p)
U Voelker, A Voelker, W G Haldenwang
The yeast two-hybrid system detects interactions between Bacillus subtilis sigmaB regulators.
J Bacteriol: 1996, 178(23);7020-3
[PubMed:8955331]
[WorldCat.org]
[DOI]
(P p)
X Yang, C M Kang, M S Brody, C W Price
Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor.
Genes Dev: 1996, 10(18);2265-75
[PubMed:8824586]
[WorldCat.org]
[DOI]
(P p)
U Voelker, A Voelker, W G Haldenwang
Reactivation of the Bacillus subtilis anti-sigma B antagonist, RsbV, by stress- or starvation-induced phosphatase activities.
J Bacteriol: 1996, 178(18);5456-63
[PubMed:8808936]
[WorldCat.org]
[DOI]
(P p)
C M Kang, M S Brody, S Akbar, X Yang, C W Price
Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor sigma(b) in response to environmental stress.
J Bacteriol: 1996, 178(13);3846-53
[PubMed:8682789]
[WorldCat.org]
[DOI]
(P p)
A Dufour, U Voelker, A Voelker, W G Haldenwang
Relative levels and fractionation properties of Bacillus subtilis σ(B) and its regulators during balanced growth and stress.
J Bacteriol: 1996, 178(13);3701-9 sigma
[PubMed:8682769]
[WorldCat.org]
[DOI]
(P p)
A A Wise, C W Price
Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor sigma B in response to environmental signals.
J Bacteriol: 1995, 177(1);123-33
[PubMed:8002610]
[WorldCat.org]
[DOI]
(P p)