Difference between revisions of "UgtP"
(→Expression and regulation) |
|||
| Line 57: | Line 57: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | + | * cells are bent and distended {{PubMed|22362028}} | |
| − | + | * increased expression of the [[SigM]], [[SigV]], and [[SigX]] regulons {{PubMed|22362028}} | |
| − | + | * altered [[localization]] of [[MreB]] (irregular clusters instead of helical dots) {{PubMed|22362028}} | |
| + | * the inactivation of ''[[ugtP]]'' suppresses the poor and filametous growth of the ''[[yvcL]] [[zapA]]'' double mutant {{PubMed|24097947}} | ||
=== Database entries === | === Database entries === | ||
| Line 160: | Line 161: | ||
<pubmed> 19680248 17662935 22575476 </pubmed> | <pubmed> 19680248 17662935 22575476 </pubmed> | ||
==Original Publications== | ==Original Publications== | ||
| − | <pubmed>9244290,18820022,17662947,9720862 22362028 15640167 23935518 22931116</pubmed> | + | <pubmed>9244290,18820022,17662947,9720862 22362028 15640167 23935518 22931116 24097947</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 18:16, 21 December 2013
- Description: UDP-glucose diacylglycerol glucosyltransferase, growth-rate dependent inhibitor of cell division
| Gene name | ugtP |
| Synonyms | ypfP |
| Essential | no |
| Product | UDP-glucose diacylglycerol glucosyltransferase |
| Function | synthesis of glycolipids and anchoring of lipoteichoic acid, inhibition of FtsZ assembly |
| Gene expression levels in SubtiExpress: ugtP | |
| Interactions involving this protein in SubtInteract: UgtP | |
| Metabolic function and regulation of this protein in SubtiPathways: Lipid synthesis | |
| MW, pI | 43 kDa, 8.398 |
| Gene length, protein length | 1146 bp, 382 aa |
| Immediate neighbours | metA, cspD |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 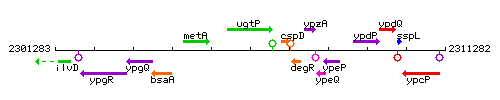
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell division, lipid metabolism/ other, cell envelope stress proteins (controlled by SigM, V, W, X, Y), membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU21920
Phenotypes of a mutant
- cells are bent and distended PubMed
- increased expression of the SigM, SigV, and SigX regulons PubMed
- altered localization of MreB (irregular clusters instead of helical dots) PubMed
- the inactivation of ugtP suppresses the poor and filametous growth of the yvcL zapA double mutant PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- UDP-glucose + 1,2-diacylglycerol = UDP + 1,2-diacyl-3-(O-beta-D-glucopyranosyl)-sn-glycerol (according to Swiss-Prot)
- the interaction with FtsZ results in inhibition of cell division and an increase of cell size PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization:
- membrane-bound protein, self-assembles into tightly wound spirals in vitro PubMed
- under nutrient rich conditions (increased concentration of UDP-Glc): throughout the cell, concentrated at the cell poles and/or the cytokinetic ring, interaction with FtsZ PubMed
- under nutrient poor conditions: forms punctate foci (oligomers), no interaction with FtsZ PubMed
Database entries
- Structure:
- UniProt: P54166
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- GP1369 (ugtP::spc), available in Jörg Stülke's lab
- Expression vector:
- pGP2571, for expression in B. subtilis (based on pBQ200, available in Jörg Stülke's lab
- pGP2600, for expression/ purification from E. coli with N-terminal His-tag, in pWH844, available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original Publications