Difference between revisions of "BsrB"
(→Biological materials) |
|||
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || antagonist of [[SigA]] | |style="background:#ABCDEF;" align="center"|'''Function''' || antagonist of [[SigA]] | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this RNA in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this RNA in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=BsrB BsrB] |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Gene length''' || 203 bp | |style="background:#ABCDEF;" align="center"| '''Gene length''' || 203 bp | ||
Revision as of 14:12, 18 November 2013
- Description: 6S RNA
| Gene name | bsrB |
| Synonyms | |
| Essential | no |
| Product | 6S RNA |
| Function | antagonist of SigA |
| Interactions involving this RNA in SubtInteract: BsrB | |
| Gene length | 203 bp |
| Immediate neighbours | yocI, azoR1 |
| Gene sequence (+200bp) | |
Genetic context 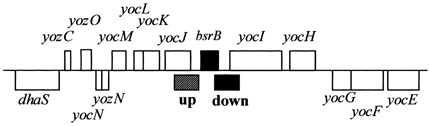
| |
Contents
Categories containing this gene/protein
sigma factors and their control, ncRNA
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU_misc_RNA_32
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: no entry
- Structure:
Additional information
- Similar RNA: bsrA
Expression and regulation
- Operon: bsrB PubMed
- Regulation: expressed during vegetative growthPubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
Labs working on this gene/RNA
- Kouji Nakamura, Tsukuba, University, Japan
- Roland Hartmann, Philipps-Universität Marburg, Marburg, Germany, homepage
Your additional remarks
References