Difference between revisions of "Des"
(→References) |
|||
| Line 152: | Line 152: | ||
=References= | =References= | ||
| − | + | == Reviews == | |
| − | <pubmed>19164152,11251847,12399512,9555904 10559169 11285232 , 12207704 12730185 19820084 12207704 20581210</pubmed> | + | <pubmed> 19363032 </pubmed> |
| + | == Original publications == | ||
| + | <pubmed>19164152,11251847,12399512,9555904 10559169 11285232 , 12207704 12730185 19820084 12207704 20581210 22416764,18154726, 21665975</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:39, 31 July 2013
- Description: phospholipid desaturase
| Gene name | des |
| Synonyms | yocE |
| Essential | no |
| Product | phospholipid desaturase |
| Function | adaptation of membrane fluidity at low temperatures |
| Gene expression levels in SubtiExpress: des | |
| Interactions involving this protein in SubtInteract: Des | |
| Metabolic function and regulation of this protein in SubtiPathways: Fatty acid degradation | |
| MW, pI | 40 kDa, 9.88 |
| Gene length, protein length | 1056 bp, 352 aa |
| Immediate neighbours | yocD, desK |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 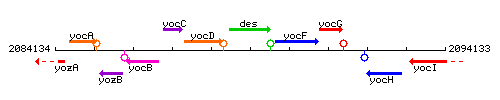
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed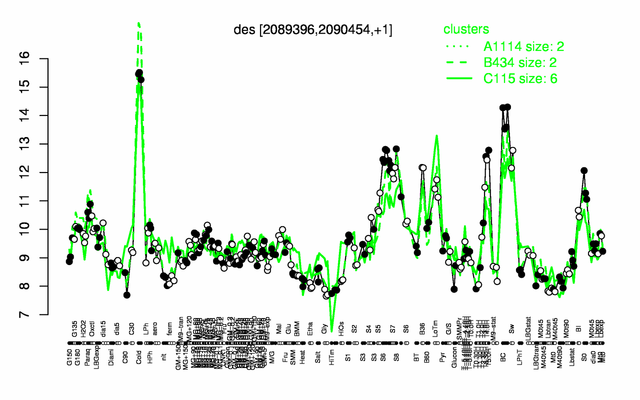
| |
Contents
Categories containing this gene/protein
lipid metabolism/ other, cold stress proteins, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU19180
Phenotypes of a mutant
- cold-sensitive PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- in strains evolved for growth at low pressure, des expression is significantly increased as compared to the wild type, this allows growth at low pressure PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: introduces double bonds into long chain fatty acids PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: integral membrane protein PubMed
Database entries
- Structure:
- UniProt: O34653
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: des PubMed
- Additional information:
- in strains evolved for growth at low pressure, des expression is significantly increased as compared to the wild type PubMed
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Diego de Mendoza, Universidad Nacional de Rosario, Argentine homepage
Your additional remarks
References
Reviews
Original publications