Difference between revisions of "MreD"
| Line 37: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 99: | Line 95: | ||
** part of the [[cell wall biosynthetic complex]] {{PubMed|21636744,21636745}} | ** part of the [[cell wall biosynthetic complex]] {{PubMed|21636744,21636745}} | ||
** [[MreD]] is a member of a suspected group of hubs proteins that were suggested to be involved in a large number of [[SubtInteract|interactions]] {{PubMed|21630458}} | ** [[MreD]] is a member of a suspected group of hubs proteins that were suggested to be involved in a large number of [[SubtInteract|interactions]] {{PubMed|21630458}} | ||
| + | ** [[RodZ]]-[[MreD]] {{PubMed|23879732}} | ||
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
| Line 126: | Line 123: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=mreD_2859317_2859835_-1 mreD] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=mreD_2859317_2859835_-1 mreD] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
** ''[[radC]]'': [[SigM]] {{PubMed|18179421}} | ** ''[[radC]]'': [[SigM]] {{PubMed|18179421}} | ||
| Line 159: | Line 156: | ||
<pubmed>, 20566861,21636744,21636745 </pubmed> | <pubmed>, 20566861,21636744,21636745 </pubmed> | ||
==Other original publications== | ==Other original publications== | ||
| − | <pubmed>1400224,18156271,16101995,19643765 8459776,18179421 21630458</pubmed> | + | <pubmed>1400224,18156271,16101995,19643765 8459776,18179421 21630458 23879732</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:20, 30 July 2013
- Description: MreD is a cell shape determining protein, it couples the cytosolic MreB and MreB-like proteins to the extracellular peptidoglycan-synthesizing machinery
| Gene name | mreD |
| Synonyms | rodB |
| Essential | yes |
| Product | cell shape-determining protein |
| Function | cell shape determation |
| Gene expression levels in SubtiExpress: mreD | |
| Interactions involving this protein in SubtInteract: MreD | |
| MW, pI | 19 kDa, 7.906 |
| Gene length, protein length | 516 bp, 172 aa |
| Immediate neighbours | minC, mreC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 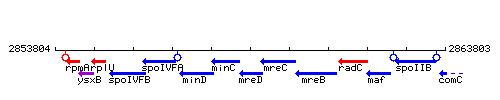
This image was kindly provided by SubtiList
| |
| Expression at a glance PubMed 500px | |
Contents
Categories containing this gene/protein
cell shape, cell envelope stress proteins (controlled by SigM, V, W, X, Y), essential genes, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU28010
Phenotypes of a mutant
- the phenotype of mreD is similar to that of mreC
- mreD is essential under normal conditions PubMed
- Depletion of MreD leads to a progressive increase in the width and a decrease in the length of the cell and cells become lytic. In the depletion strain, lysis can be prevented and cell growth, but not cell shape, can be recovered by inculaion of Magnesium in the media. This shape defect is consistent with a role for mreD in cell wall synthesis during elongation and has a similar phenotype to other genes with roles in elongation.
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- MreD functions in cell wall synthesis by, together with the MreB cytoskeleton, localizing the cell wall synthetic machinery to the correct part of the cell. As a transmembrane protein MreD is thought to provide a patch on the membrane that MreB inteacts with. MreC therefore ensures that the cell wall is made in the correct way to maintain the proper shape of the cell.
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: None/ structural
- Protein family: mreD family (according to Swiss-Prot) COG2891
- Paralogous protein(s): None
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- part of the cell wall biosynthetic complex PubMed
- MreD is a member of a suspected group of hubs proteins that were suggested to be involved in a large number of interactions PubMed
- RodZ-MreD PubMed
- Localization:
- trans-membrane protein PubMed
- during logarithmic growth, MreD forms discrete patches thst move processively along peripheral tracks perpendicular to the cell axis PubMed
- forms transverse bands as cells enter the stationary phase PubMed
- reports on helical structures formed by MreD PubMed seem to be misinterpretation of data PubMed
Database entries
- Structure: None
- UniProt: Q01467
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: A conditional mutant with an inframe deletion of mreD complemented by a xylose inducible copy at an ectopic locus (strain named 4352) is avaliable from the Errington Group.
- Expression vector:
- lacZ fusion:
- GFP fusion: A functional N-terminal GFP fusion has been made where the fusion protein is the only copy of the gene in the cell: strain 3416 PubMed.
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Peter Graumann, Freiburg University, Germany homepage
Your additional remarks
References
Localization
Other original publications
Katarína Muchová, Zuzana Chromiková, Imrich Barák
Control of Bacillus subtilis cell shape by RodZ.
Environ Microbiol: 2013, 15(12);3259-71
[PubMed:23879732]
[WorldCat.org]
[DOI]
(I p)
Elodie Marchadier, Rut Carballido-López, Sophie Brinster, Céline Fabret, Peggy Mervelet, Philippe Bessières, Marie-Françoise Noirot-Gros, Vincent Fromion, Philippe Noirot
An expanded protein-protein interaction network in Bacillus subtilis reveals a group of hubs: Exploration by an integrative approach.
Proteomics: 2011, 11(15);2981-91
[PubMed:21630458]
[WorldCat.org]
[DOI]
(I p)
Kathrin Schirner, Jeff Errington
Influence of heterologous MreB proteins on cell morphology of Bacillus subtilis.
Microbiology (Reading): 2009, 155(Pt 11);3611-3621
[PubMed:19643765]
[WorldCat.org]
[DOI]
(I p)
Warawan Eiamphungporn, John D Helmann
The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses.
Mol Microbiol: 2008, 67(4);830-48
[PubMed:18179421]
[WorldCat.org]
[DOI]
(P p)
Alex Formstone, Rut Carballido-López, Philippe Noirot, Jeffery Errington, Dirk-Jan Scheffers
Localization and interactions of teichoic acid synthetic enzymes in Bacillus subtilis.
J Bacteriol: 2008, 190(5);1812-21
[PubMed:18156271]
[WorldCat.org]
[DOI]
(I p)
Mark Leaver, Jeff Errington
Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis.
Mol Microbiol: 2005, 57(5);1196-209
[PubMed:16101995]
[WorldCat.org]
[DOI]
(P p)
S Lee, C W Price
The minCD locus of Bacillus subtilis lacks the minE determinant that provides topological specificity to cell division.
Mol Microbiol: 1993, 7(4);601-10
[PubMed:8459776]
[WorldCat.org]
[DOI]
(P p)
P A Levin, P S Margolis, P Setlow, R Losick, D Sun
Identification of Bacillus subtilis genes for septum placement and shape determination.
J Bacteriol: 1992, 174(21);6717-28
[PubMed:1400224]
[WorldCat.org]
[DOI]
(P p)