Difference between revisions of "MutTA"
| Line 27: | Line 27: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| − | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=ytkD_3134983_3135459_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:mutTA_expression.png|500px]] | + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=ytkD_3134983_3135459_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:mutTA_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU30630]] |
|- | |- | ||
|} | |} | ||
Revision as of 14:22, 16 May 2013
- Description: error prevention oxidized guanine system, confers protection against oxidative stress to vegetative cells, moreover the protein has RNA pyrophosphohydrolase activity
| Gene name | mutTA |
| Synonyms | ytkD, rppH |
| Essential | no |
| Product | 8-oxo-dGTPase (antimutator), RNA pyrophosphohydrolase |
| Function | DNA repair, RNA degradation |
| Gene expression levels in SubtiExpress: mutTA | |
| MW, pI | 18 kDa, 5.952 |
| Gene length, protein length | 474 bp, 158 aa |
| Immediate neighbours | ytlD, ytkC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed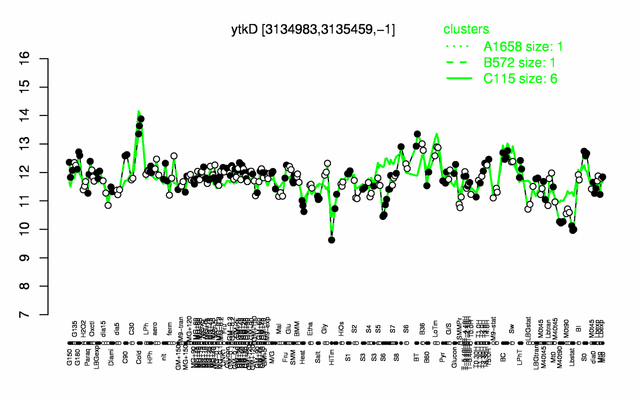
| |
Contents
Categories containing this gene/protein
DNA repair/ recombination, sporulation proteins, RNases
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU30630
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- removal of pyrophosphate from the 5' end of mRNAs to make the RNA accessible for degradation by RNases PubMed
- RppH requires at least two unpaired nucleotides at the 5' end of its RNA substrates and prefers three or more. The second of these 5'-terminal nucleotides must be G, whereas a less strict preference for a purine is evident at the third position, and A is slightly favored over G at the first position PubMed
- Protein family: Nudix hydrolase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: O35013
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Ping-kun Hsieh, Jamie Richards, Quansheng Liu, Joel G Belasco
Specificity of RppH-dependent RNA degradation in Bacillus subtilis.
Proc Natl Acad Sci U S A: 2013, 110(22);8864-9
[PubMed:23610425]
[WorldCat.org]
[DOI]
(I p)
Jérémie Piton, Valéry Larue, Yann Thillier, Audrey Dorléans, Olivier Pellegrini, Inés Li de la Sierra-Gallay, Jean-Jacques Vasseur, Françoise Debart, Carine Tisné, Ciarán Condon
Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc Natl Acad Sci U S A: 2013, 110(22);8858-63
[PubMed:23610407]
[WorldCat.org]
[DOI]
(I p)
Jamie Richards, Quansheng Liu, Olivier Pellegrini, Helena Celesnik, Shiyi Yao, David H Bechhofer, Ciarán Condon, Joel G Belasco
An RNA pyrophosphohydrolase triggers 5'-exonucleolytic degradation of mRNA in Bacillus subtilis.
Mol Cell: 2011, 43(6);940-9
[PubMed:21925382]
[WorldCat.org]
[DOI]
(I p)
Luz E Vidales, Lluvia C Cárdenas, Eduardo Robleto, Ronald E Yasbin, Mario Pedraza-Reyes
Defects in the error prevention oxidized guanine system potentiate stationary-phase mutagenesis in Bacillus subtilis.
J Bacteriol: 2009, 191(2);506-13
[PubMed:19011023]
[WorldCat.org]
[DOI]
(I p)
Francisco X Castellanos-Juárez, Carlos Alvarez-Alvarez, Ronald E Yasbin, Barbara Setlow, Peter Setlow, Mario Pedraza-Reyes
YtkD and MutT protect vegetative cells but not spores of Bacillus subtilis from oxidative stress.
J Bacteriol: 2006, 188(6);2285-9
[PubMed:16513759]
[WorldCat.org]
[DOI]
(P p)
Stephanie T Wang, Barbara Setlow, Erin M Conlon, Jessica L Lyon, Daisuke Imamura, Tsutomu Sato, Peter Setlow, Richard Losick, Patrick Eichenberger
The forespore line of gene expression in Bacillus subtilis.
J Mol Biol: 2006, 358(1);16-37
[PubMed:16497325]
[WorldCat.org]
[DOI]
(P p)
Wenlian Xu, Candice R Jones, Christopher A Dunn, Maurice J Bessman
Gene ytkD of Bacillus subtilis encodes an atypical nucleoside triphosphatase member of the Nudix hydrolase superfamily.
J Bacteriol: 2004, 186(24);8380-4
[PubMed:15576788]
[WorldCat.org]
[DOI]
(P p)
Martha I Ramírez, Francisco X Castellanos-Juárez, Ronald E Yasbin, Mario Pedraza-Reyes
The ytkD (mutTA) gene of Bacillus subtilis encodes a functional antimutator 8-Oxo-(dGTP/GTP)ase and is under dual control of sigma A and sigma F RNA polymerases.
J Bacteriol: 2004, 186(4);1050-9
[PubMed:14761999]
[WorldCat.org]
[DOI]
(P p)
Alia Lapidus, Nathalie Galleron, Alexei Sorokin, S Dusko Ehrlich
Sequencing and functional annotation of the Bacillus subtilis genes in the 200 kb rrnB-dnaB region.
Microbiology (Reading): 1997, 143 ( Pt 11);3431-3441
[PubMed:9387221]
[WorldCat.org]
[DOI]
(P p)