Difference between revisions of "LytG"
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || cell wall turnover | |style="background:#ABCDEF;" align="center"|'''Function''' || cell wall turnover | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU31120 lytG] |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 31 kDa, 8.277 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 31 kDa, 8.277 | ||
| Line 22: | Line 22: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yubF]]'', ''[[yubD]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yubF]]'', ''[[yubD]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU31120 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU31120 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU31120 Advanced_DNA] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:yubE_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:yubE_context.gif]] | ||
Revision as of 13:42, 13 May 2013
- Description: similar to N-acetylmuramoyl-L-alanine amidase
| Gene name | lytG |
| Synonyms | yubE |
| Essential | no |
| Product | unknown |
| Function | cell wall turnover |
| Gene expression levels in SubtiExpress: lytG | |
| MW, pI | 31 kDa, 8.277 |
| Gene length, protein length | 846 bp, 282 aa |
| Immediate neighbours | yubF, yubD |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 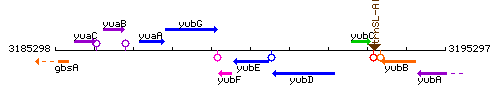
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed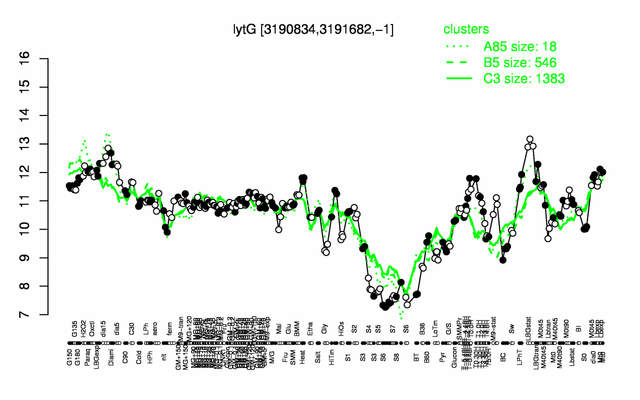
| |
Contents
Categories containing this gene/protein
cell wall degradation/ turnover
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU31120
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Hydrolysis of 1,4-beta-linkages between N-acetyl-D-glucosamine and N-acetylmuramic acid residues in a peptidoglycan (according to Swiss-Prot)
- Protein family: glycosyl hydrolase 73 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- secreted (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: O32083
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Fusinita van den Ent, Mark Leaver, Felipe Bendezu, Jeff Errington, Piet de Boer, Jan Löwe
Dimeric structure of the cell shape protein MreC and its functional implications.
Mol Microbiol: 2006, 62(6);1631-42
[PubMed:17427287]
[WorldCat.org]
[DOI]
(P p)
Gavin J Horsburgh, Abdelmadjid Atrih, Michael P Williamson, Simon J Foster
LytG of Bacillus subtilis is a novel peptidoglycan hydrolase: the major active glucosaminidase.
Biochemistry: 2003, 42(2);257-64
[PubMed:12525152]
[WorldCat.org]
[DOI]
(P p)