Difference between revisions of "Pyk"
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || catabolic enzyme in glycolysis | |style="background:#ABCDEF;" align="center"|'''Function''' || catabolic enzyme in glycolysis | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU29180 pyk] |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism]''' | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism]''' | ||
| Line 24: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ytzA]]'', ''[[pfkA]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ytzA]]'', ''[[pfkA]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU29180 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU29180 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU29180 Advanced_DNA] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:pyk_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:pyk_context.gif]] | ||
Revision as of 13:34, 13 May 2013
- Description: pyruvate kinase, glycolytic enzyme
| Gene name | pyk |
| Synonyms | pykA |
| Essential | no |
| Product | pyruvate kinase |
| Function | catabolic enzyme in glycolysis |
| Gene expression levels in SubtiExpress: pyk | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 61,9 kDa, 4.88 |
| Gene length, protein length | 1755 bp, 585 amino acids |
| Immediate neighbours | ytzA, pfkA |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 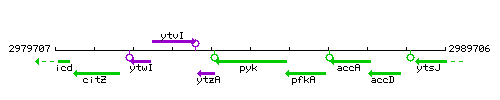
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
ATP synthesis, carbon core metabolism, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29180
Phenotypes of a mutant
Unable to grow with non-PTS carbohydrates (such as glucitol or glycerol) as single carbon source.
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ADP + phosphoenolpyruvate --> ATP + pyruvate
- The reaction is irreversible under physiological conditions
- Protein family: PEP-utilizing enzyme family (according to Swiss-Prot) pyruvate kinase family, (C-terminal section: PEP-utilizing enzyme family)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information: Allosteric Regulation PubMed
- Domains:
- Modification: phosphorylation on Ser36 PubMed, PubMed, phosphorylation on Ser536 or Ser546 PubMed, please note that the Ser is not on position 536 but rather at 538
- Cofactor(s): Mg2+, K+
- Effectors of protein activity:
- Localization: cytoplasm PubMed
Database entries
- Structure: 2E28 (Geobacillus stearothermophilus)
- UniProt: P80885
- KEGG entry: [3]
- E.C. number: 2.7.1.40
Additional information
The enzyme is a tetramer with four active sites PubMed
Expression and regulation
- Sigma factor:
- Regulation:
- twofold induced by glucose PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Expression vector:
- expression in E. coli, N-terminal His-tag: pGP1100 (in pWH844), available in Stülke lab
- expression in B. subtilis, native protein: pGP1411 (in pBQ200), available in Stülke lab
- expression in B. subtilis, N-terminal Strep-tag: pGP1409 (in pGP380), available in Stülke lab
- expression in B. subtilis, C-terminal Strep-tag: pGP1410 (in pGP382), available in Stülke lab
- lacZ fusion: see pfkA
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
Natividad Cabrera-Valladares, Luz M Martínez, Noemí Flores, Georgina Hernández-Chávez, Alfredo Martínez, Francisco Bolívar, Guillermo Gosset
Physiologic consequences of glucose transport and phosphoenolpyruvate node modifications in Bacillus subtilis 168.
J Mol Microbiol Biotechnol: 2012, 22(3);177-97
[PubMed:22846916]
[WorldCat.org]
[DOI]
(I p)
Christopher T Brown, Laura K Fishwick, Binna M Chokshi, Marissa A Cuff, Jay M Jackson, Travis Oglesby, Alison T Rioux, Enrique Rodriguez, Gregory S Stupp, Austin H Trupp, James S Woollcombe-Clarke, Tracy N Wright, William J Zaragoza, Jennifer C Drew, Eric W Triplett, Wayne L Nicholson
Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation.
Appl Environ Microbiol: 2011, 77(19);6867-77
[PubMed:21821766]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Dörte Becher, Jörg Bernhardt, Katrin Gronau, Anja Klutzny, Michael Hecker
Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis.
Proteomics: 2007, 7(19);3509-26
[PubMed:17726680]
[WorldCat.org]
[DOI]
(P p)
Laurent Jannière, Danielle Canceill, Catherine Suski, Sophie Kanga, Bérengère Dalmais, Roxane Lestini, Anne-Françoise Monnier, Jérôme Chapuis, Alexander Bolotin, Marina Titok, Emmanuelle Le Chatelier, S Dusko Ehrlich
Genetic evidence for a link between glycolysis and DNA replication.
PLoS One: 2007, 2(5);e447
[PubMed:17505547]
[WorldCat.org]
[DOI]
(I e)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Alain Lévine, Françoise Vannier, Cédric Absalon, Lauriane Kuhn, Peter Jackson, Elaine Scrivener, Valérie Labas, Joëlle Vinh, Patrick Courtney, Jérôme Garin, Simone J Séror
Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes.
Proteomics: 2006, 6(7);2157-73
[PubMed:16493705]
[WorldCat.org]
[DOI]
(P p)
H Ludwig, G Homuth, M Schmalisch, F M Dyka, M Hecker, J Stülke
Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon.
Mol Microbiol: 2001, 41(2);409-22
[PubMed:11489127]
[WorldCat.org]
[DOI]
(P p)
B Fry, T Zhu, M M Domach, R R Koepsel, C Phalakornkule, M M Ataai
Characterization of growth and acid formation in a Bacillus subtilis pyruvate kinase mutant.
Appl Environ Microbiol: 2000, 66(9);4045-9
[PubMed:10966427]
[WorldCat.org]
[DOI]
(P p)
H Sakai, K Suzuki, K Imahori
Purification and properties of pyruvate kinase from Bacillus stearothermophilus.
J Biochem: 1986, 99(4);1157-67
[PubMed:3711058]
[WorldCat.org]
[DOI]
(P p)
M Diesterhaft, E Freese
Pyruvate kinase of bacillus subtilis.
Biochim Biophys Acta: 1972, 268(2);373-80
[PubMed:4623707]
[WorldCat.org]
[DOI]
(P p)